Figure 3. Motif segregation.
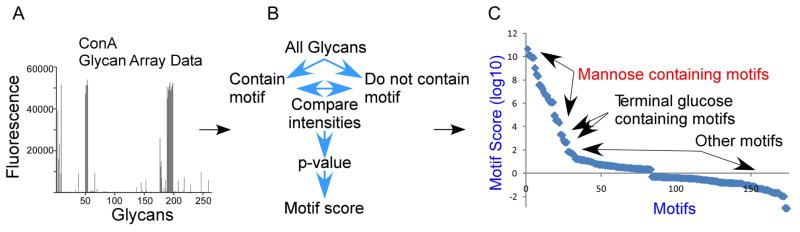
Quantified glycan array data (A) are processed using the GlycoSearch software and the motif segregation algorithm to produce motif scores for each experiment (B). The analysis of a glycan array experiment of the lectin ConA is given as an example, in which the top scoring motifs contain mannose, followed by motifs with terminal glucose (C).
