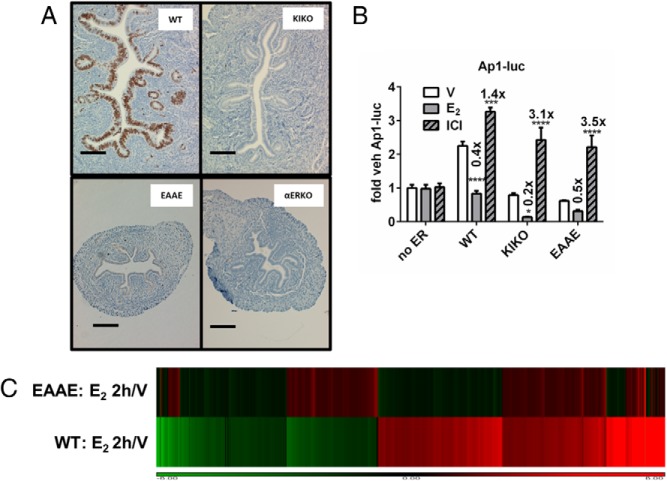
Figure 6.
Evaluation of DBD mutant ERα activities in vivo and in vitro. A, Uterine cross sections from ovariectomized WT, KIKO, EAAE, or ERα-null mice that were treated for 24 hours with E2. Sections were evaluated for the proliferative marker, Ki67 (brown). Scale bar corresponds to 0.1 mm. B, WT, KIKO, and EAAE ERα mediate AP1-luc responses similarly. WT, KIKO, and EAAE ERα with AP1-luc. Fold changes relative to vehicle (V) treatment are indicated above each bar. E2, 10 nM; ICI 182,780 (ICI), 100 nM. Statistical analysis included 2-way ANOVA, multiple comparisons of means vs results for vehicle treatment, and Bonferroni multiple test correction. ***, P < .001; ****, P < .0001. C, Microarray profile. A hierarchical cluster of normalized ratios (E2 2h/V) of WT and EAAE uterine RNA is shown. Uterine RNA from WT or EAAE ovariectomized mice collected 2 hours after vehicle (V) or E2 injection (n = 3 each condition) was compared by microarray. The cluster was created by combining replicates and building ratios (E2 treated/vehicle treated) and filtering for probes that met the following criteria: absolute fold change of > 2 in at least 1 genotype, probe intensity of >100 in at least 1 condition, and false discovery rate < 0.05. This resulted in 3162 probes. Red indicates induced transcripts; green signifies repression.