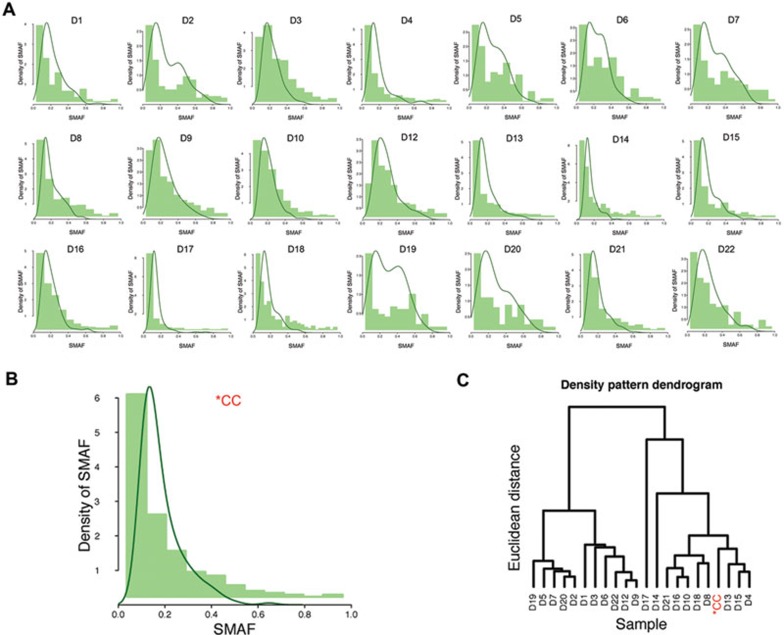
Figure 4.
Heterogeneity of clonal origin in colon cancer. (A) The density distribution of SMAFS of 21 whole-tissue exome-sequenced colon cancer was analyzed by kernel density estimate (KDE). Tumors of different clonality were expected to give rise to different KDE patterns. (B) KDE plot of the same tissue specimen (*CC) that was subject to single-cell sequencing was extracted from whole-tissue exome-sequencing data. Similar to SMAFS derived from single-cell sequencing, this specimen exhibits a peak in the frequency window of 20%-30% in the KDE plot of whole-tissue exome-sequencing data. (C) Hierarchical clustering was performed to segregate colon cancer samples into subgroups based on KDE patterns. It is inferred that specimens D4, D13 and D15 clustered along with *CC are of possible biclonal origin.