Abstract
The neuropeptide gonadotropin-releasing hormone stimulates synthesis and secretion of the glycoprotein gonadotropic hormones and activates the unfolded protein response, which causes a transient reduction of endoplasmic reticulum-associated mRNA translation. Hormone-treated cell extracts were fractionated to resolve mRNA in active polyribosomes from mRNA in inactive complexes. Quantitative real-time PCR and expression array analysis were used to determine hormone-induced redistribution of mRNAs between fractions and individual mRNAs were found to be redistributed differentially. Among the affected mRNAs relevant to gonadotropin synthesis, the luteinizing hormone subunit genes Lhb and Cga were enriched in the ribonucleoprotein pool. The MAP kinase phosphatase Dusp1 was enriched in the polyribosome pool. Enrichment of Dusp1 mRNA in the polyribosome pool was independent of the unfolded protein response, sensitive to ERK inhibition, and dependent on the 3′ untranslated region. The results show that GnRH exerts translational control to modulate physiologically relevant gene expression through two distinct signaling pathways.
Keywords: Gonadotropins, Luteinizing Hormone, Dusp1, Pituitary, Translation, Unfolded Protein Response
1. Introduction
The reproductive endocrine axis is controlled by the timed release of gonadotropin-releasing hormone I (GnRH) from hormone-producing neurons of the hypothalamus into the fenestrated capillary bed providing arterial flow to the anterior pituitary. Timing and amplitude of GnRH pulses alters production and secretion of the gonadotropins Luteinizing Hormone (LH) and Follicle-Stimulating Hormone (FSH) by the gonadotropes. Production of LH is favored by high frequency, high amplitude pulses of GnRH whereas FSH is favored by lower pulse frequencies (Ferris and Shupnik, 2006; Tsaneva-Atanasova et al., 2012). The altered production and timing of gonadotropin release in response to GnRH stimulation is essential for the onset of the preovulatory LH surge in females. The gonadotropins are heterodimeric glycoprotein hormones comprised of a common alpha subunit encoded by the Cga gene, and a unique beta subunit encoded by the Lhb and Fshb genes for LH and FSH, respectively. The transcriptional control of the gonadotropin genes has been extensively studied both in vivo (Burger et al., 2004; Burger et al., 2011) and using in vitro cell models such as LβT2 cells derived from LH-producing cells of the mouse anterior pituitary (Alarid et al., 1996), or modified cell lines engineered to express the GnRH receptor (Armstrong et al., 2009).
Both basal and GnRH pulse-induced transcriptional regulation of the gonadotropin genes have been well characterized. Regulation of gene expression by GnRH is largely, but not exclusively modulated by signaling via the mitogen-activated protein kinase (MAP kinase) cascades, particularly through activation of the extracellular receptor signal activated kinases MAPK 1 and 3, which target transcription through activation of immediate-early genes and increase protein synthesis via cap-dependent translation initiation (Lawson et al., 2007; Nguyen et al., 2004). The MAP kinase signaling cascade experiences rapid negative feedback which is partly mediated by the dual specificity protein phosphatases including DUSP1, and this is hypothesized to contribute to the overall ability of the gonadotrope to interpret GnRH pulse frequency and amplitude (Lim et al., 2009). The expression of Dusp1 and related dual specificity kinase family members is induced by pulsatile GnRH (Lawson et al., 2007). DUSP1 protein synthesis is also induced by GnRH, as is its rapid phosphorylation. Manipulation of DUSP1 levels alters MAPK 1/3 phosphorylation and the Egr1 and Lhb transcriptional response to GnRH stimulation (Caunt et al., 2008; Nguyen et al., 2010; Zhang and Roberson, 2006). Although the MAP kinases and the DUSP’s comprise an ultra-short feedback loop that provides rapid but transient propagation of GnRH –induced signaling events to maintain sensitivity to subsequent GnRH pulses, there are compelling findings in HeLa cells that suggest this may not fully explain pulse sensitivity (Armstrong et al., 2010). The contribution of rapid transcriptional activation by EGR1 and rapid signaling feedback via DUSP1 represent two distinct points of control that can potentially facilitate rapid response and resolution to GnRH receptor signaling events.
Another regulatory cascade initiated by GnRH treatment is the unfolded protein response, or UPR (Do et al., 2009). This pathway involves both transcriptional and protein synthesis regulatory arms that serve to minimize endoplasmic reticulum (ER) stress. In secretory cells, stress occurs after a secretion event due to loss of calcium ion from the ER lumen and alteration of the oxidative environment. The UPR is essential to preserve normal function of pancreatic β-cells, hepatocytes, osteoblasts, and plasma cells, and is generally regarded as an indispensable control mechanism in cells experiencing high secretory demand (Scheuner and Kaufman, 2008; Zhang and Kaufman, 2006). Proper folding and export of proteins destined for secretion or targeted to extracellular or intracellular membranes through the ER requires a proper oxidative environment. Disruption of the ER luminal environment leads to accumulation of improperly folded protein that can lead to reduced ER capacity and cell death (Fribley et al., 2009; Walter and Ron, 2011). Protection of ER balance and maintenance of protein quality is controlled by the UPR, which blocks the import of protein into the ER, degrades unfolded proteins, and induces an adaptive response to increase ER capacity. Pathological conditions such as hypoxia, inflammation, oxidative stress, or chemical insult can also lead to induction of the UPR.
We have shown that GnRH induces the UPR in gonadotropes and in the cultured gonadotrope cell line LβT2 (Do et al., 2009). Induction of the UPR is transient in LβT2 cells, resolving within 60 minutes. A hallmark of translational pausing is activation of the ER membrane-resident eukaryotic initiation factor 2 alpha kinase 3 (EIF2AK3). Activation of EIF2AK3 results in phosphorylation and inactivation of cytosolic eukaryotic initiation factor 2 alpha (EIF2A), an essential part of the translation initiation complex. This results in inhibition of translation and subsequent protein export into the ER lumen. Examination of LβT2 cell polyribosome profiles by centrifugation on sucrose density gradients showed that after GnRH treatment there is a transient redistribution of ribosomes between active and inactive states as measured by RNA content in active, polyribosome fractions versus inactive ribonucleoprotein (RNP) complexes. Examination of Cga and Lhb mRNAs across polyribosome and RNP fractions showed that both were transiently displaced from the polyribosome to the RNP fractions by GnRH, consistent with translational pausing.
It is not known if pausing is a generalized response to UPR induction, or if gonadotropin mRNAs represent a subpopulation of mRNAs targeted for translational pausing. We examined the details of polyribosome redistribution induced by GnRH treatment to assess the specificity of translational pausing induced by GnRH in LβT2 cells. Using a microarray-based approach to evaluate mRNA content in both polyribosome and RNP fractions and specific analysis of individual mRNA species, we find that mRNAs are differentially enriched or redistributed to both the polyribosome and RNP fractions after GnRH treatment. Evaluation of the signaling components participating in translational control show that redistribution to the polyribosome fractions is independent of EIF2AK3 activation and may represent an independent mRNA regulatory process.
2. Materials and Methods
2.1. Cell Culture
The LβT2 mouse gonadotrope line (Alarid et al., 1996) was maintained in high-glucose (4.5 g/L) 20 mM HEPES-buffered DMEM supplemented with penicillin/streptomycin and 10% fetal bovine serum. The cells were maintained at 37° C in a humidified atmosphere of 5% CO2. Prior to GnRH treatment, cells were changed into serum-free DMEM for 12–16 hours. For transfection studies, cells were transfected immediately after changing to serum-free medium and allowed to incubate 12–16 hours before GnRH treatments.
2.2. Plasmid construction and transfection
To analyze the effect of the 3′ UTR of Dusp1 or Lhb, 3′ UTR on reporter gene expression these regions were inserted into a multiple cloning site placed downstream of the luciferase coding sequence of the pGL3-TK80C-XPL plasmid. This reporter vector was constructed by inserting the 80 bp proximal sequence of the HSV thymidine kinase promoter upstream of the luciferase coding sequence in pGL3-basic (Promega, Madison, WI) and inserting a second multiple cloning site downstream of the luciferase coding sequence prior to the SV40 termination and poly-adenylation site. The 76-bp 3′ UTR fragment of Lhb amplified by PCR from genomic DNA of C57/Bl6 mice was inserted into the EcoR1- and XhoI-digested site of pGL3-TK80C. The 688-bp 3′ UTR fragment of the Dusp1 3′UTR was amplified by PCR from genomic DNA of C57/Bl6 mice and inserted into EcoR1-digested pGL3-TK80C. These new plasmids, pTK-Luc-Lhb-3′ UTR and pTK-Luc-Dusp1 3′ UTR, were then sequenced to confirm identify.
To analyze the effect of the ELAV1 binding site in the Dusp1 3′ UTR on luciferase expression, two plasmids containing either just the previously described ELAV1 binding site (Kuwano et al., 2008) or the remainder of the 3′UTR without the site were constructed from the parent Dusp1 3′UTR plasmid. The resultant plasmids, pTK-Luc-ELAV +, and pTK-Luc-ELAV −, were sequenced to confirm identity.
For transfection, LβT2 cells were plated at a density of 2.75 × 105 cells/cm2 and incubated for 24 h before transfection. Cells were then changed into serum-free medium for 12–16 h and then transfected with Fugene 6 (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions using above plasmids and the internal control pGL3-CMV-βGal (Nguyen et al., 2004). After incubation for 16 h. the cells were treated with 10 nM GnRH (Sigma Aldrich, Inc.), or 100 nM phorbol-12-myristate-13-acetate (PMA; Calbiochem) for the indicated time. The cells were then harvested with Reporter Lysis Buffer (Promega, Madison, WI). Lysates were assayed using the Luciferase Assay Kit (Promega, Madison, WI) and Galacto-Light Plus Kit (Applied Biosystems, Foster City, CA), respectively. Luminescence was measured in a Veritas microplate luminometer (Turner BioSystem, Sunnyvale, CA).
2.3. Quantitative real-time PCR
For quantitative PCR analysis of fractionated, ribosome-bound poly-adenylated mRNA, cDNA was synthesized using Omniscript Reverse Transcriptase (Qiagen, Valencia, CA) and 10 uM of random hexamer primers (Applied Biosystems, Foster City, CA). For analysis of total RNA, cDNA was synthesized using QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA). Quantitative real-time PCR was carried out using the MyIQ Single-Color Real-Time Detection System (Bio-Rad Laboratories, Hercules, CA). The PCR products were amplified in the presence of SYBR Green using the QuantiTect SYBR Green PCR kit (Qiagen, Valencia, CA) supplemented with 300 nM of transcript specific primers and 1 μM fluorescein for proper instrument calibration. The cycling conditions used were those recommended by the PCR kit manufacturer. A melt curve was performed after each PCR run to ensure that just a single product was amplified in each assay and the size of the products were verified using agarose gel electrophoresis
Primer sequences were designed against murine mRNA sequences as available through PubMed. Sequences of the primers used were: Cga fwd, 5′-GGTTCCAAAGAATATTACCTCG-3′, Cga rev, GTCATTCTGGTCATGCTGTCC, Lhb fwd, 5′-CTGTCAACGCAACTCTGG-3′, Lhb rev, 5′-ACAGGAGGCAAAGCAGC-3′, Gapdh fwd, 5′-TGCACCACCAACTGCTTAG-3′; Gapdh rev, 5′-GATGCAGGGATGATGTTC-3′; Gnrhr fwd, 5′-GCCCCTTGCTGTACAAAGC-3′; Gnrhr rev, 5′-CCGTCTGCTAGGTAGATCATCC-3′; c-Myc fwd, 5′-GCTGCATGAGGAGACACCG-3′; c-Myc rev, 5′-CCTCGGGATGGAGATGAGC-3′; Egr1 fwd, 5′-ATTTTTCCTGAGCCCCAAAGC-3′; Egr1 rev, 5′-ATGGGAACCTGGAAACCACC-3′; Dusp1 fwd, 5′-TGGTTCAACGAGGCTATTGAC-3′; Dusp1 rev, 5′-GGCAATGAACAAACACTCTCC-3′;.
Primers were designed to generate an 80–150 bp amplicon that crossed intron/exon boundaries where possible in order to avoid amplification of any contaminating unspliced RNA or genomic DNA. All PCR reactions were performed in triplicate, except for those involving cDNA from primary pituitary cells, which were performed in duplicate. A larger cDNA fragment of all the genes assayed was inserted into pCR2.1 (Invitrogen, Carlsbad, CA) and used to generate a standard curve and assess PCR efficiency for each run. The mass values for each transcript were extrapolated from the standard curve. To calculate a fold-redistribution value for an mRNA’s movement within the ribosome profile, the mass of each transcript in each ribosome pool (polysome or RNP) was measured and a ratio was calculated for the mass of that transcript in the RNP pool compared to the polysome pool. A ratio was calculated for both the treated and control (vehicle treated) conditions. A fold-redistribution value was then generated by taking a ratio of the ratios: the RNP/polysome ratio in treated compared to the RNP/polysome ratio in control cells. Statistics were performed as indicated in each figure legend on these values. For ease of representation in the histograms only, the negative inverse was calculated and reported for fold-redistribution values between 0 and 1, such that movement of an mRNA into the polysome pool would report a negative redistribution value and movement into RNP complexes would report a positive value.
2.4. Western blotting
Protein was harvested using standard RIPA lysis buffer supplemented with phosphatase inhibitors. Standard SDS-PAGE and semi-dry transfer method (Bio-Rad Laboratories, Hercules, CA) was used to transfer the extracts onto PVDF membranes (Bio-Rad Laboratories, Hercules, CA). Blocking and primary antibody delivery was performed in 1X casein (Vector Laboratories, Burlingame, CA). Antibodies directed against phosphorylated MAPK1/3 (Santa Cruz Biotechnology, Santa Cruz, CA), total MAPK1/3 (Santa Cruz Biotechnology, Santa Cruz, CA), phosphorylated EIF2AK3 (Rockland Immunochemicals, Gilbertsville, PA), total EIF2AK3 (Rockland Immunochemicals, Gilbertsville, PA), phosphorylated eIF4E (Ser209; Cell Signaling, Beverly, MA), total eIF4E (Cell Signaling, Beverly, MA), phosphorylated 4EBP1 (Thr70; Cell Signaling, Beverly, MA), and total 4EBP1 (Cell Signaling, Beverly, MA) were incubated for 16 hours. Blots were developed using 1:2000 dilution of biotinylated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) and Chemiglow chemiluminescence (Cell Biosciences, San Leandro, CA). Chemiluminescence was visualized using the GeneSnap (Syngene, Frederick, MD) or the Flourchem Q (Cell Biosciences, San Lendro, CA) Imaging Systems.
2.5. Ribosome fractionation and mRNA isolation
Two 10-cm plates were used for each experimental condition with 2×107 LβT2 cells seeded per plate. Cells were incubated in serum free-DMEM for 16h. Cells were then incubated with Earle’s Balanced Salt Solution (EBSS) 1 hour, then treated with vehicle, 10 nM GnRH (Sigma Aldrich, St. Louis, MO) 2 mM dithiothreitol (DTT; Sigma Aldrich, St. Louis, MO.), or 100 nM phorbol-12-myristate-13-acetate (PMA; EMD Chemicals, San Diego, CA), 30 μM PD98059 (EMD Chemicals, San Diego), and 10 nM Rapamycin (Rap, EMD Chemicals, San Diego, CA) for 30 min. After treatment, 100 μg/ml cycloheximide (Sigma-Aldrich, St. Louis, MO) was added to all the plates (except for any pre-treated plates) and the cells were incubated on ice for 5 minutes. The cells were then harvested in 500 μl of ice-cold polysome extraction buffer (PEB) containing 140 mM KCl, 5 mM MgCl2, 1 mg/ml heparin sodium salt, and 20 mM Tris-HCl pH 8, supplemented with fresh 0.5 mM DTT and 100 μg/ml cycloheximide on the day of the experiment. Cells from two plates of the same experimental condition were collected and pooled at this step. The cells were spun down at 1000 X g for 3 minutes and then each pellet was resuspended in 200 μl PEB containing 1% Triton-X. The pellets were allowed to lyse on ice for 20 min with gentle inversion by hand every few minutes. Finally, the cellular debris was collected by centrifugation at 10,000 x g for 10 min. The supernatants were layered onto 5 ml 10–50% sucrose gradients made with PEB lacking Triton-X. The samples were centrifuged in a SW-55 swing-bucket rotor for 1.5 h at 150,000 X g. A 21-guage needle was used to collect fractions from the bottom of the gradient. Fractions were collected using a peristaltic pump while monitoring real-time absorption at 254 nm through a UVM-II monitor (GE Healthcare, Fairfield, CT). Fractions of 250 ul volume each were collected using a FC 203B fraction collector (Gilson, Middleton, WI). Monitoring of UV absorption and fraction collection was controlled through a Labview virtual instrument (National Instruments, Austin, TX) and a KPCI 3108 DAQ card (Keithley Instruments, Cleveland, OH). Fractions were dripped directly into SDS for a final SDS concentration of 1%. Each fraction was then treated with 0.2 mg/ml proteinase K at 37° C for 1.5 h. RNA was purified from each fraction by phenol-chloroform extraction and ethanol precipitation. An aliquot (1 ul) of RNA from each fraction was subjected to agarose electrophoresis in the presence of ethidium bromide to verify the presence of the large and small ribosomal subunits through presence of the 28S and 18S rRNAs, respectively. The rest of the RNA from the fractions was pooled into polysome and RNP pools according to the gradient’s UV absorption profile. Poly-A RNA was then isolated using Oligotex mRNA Mini Kit oligo-dT columns (Qiagen, Valencia, CA) according to the manufacturer’s recommendations and eluted into a final volume of 40 μl for each pool. A 10 μl aliquot of mRNA was used for reverse-transcription for quantitative PCR analysis.
2.6. Microarray analysis
The RNP (R) and polysome (P) fractionated mRNA collected from three independent experiments of GnRH or vehicle-treated LβT2 cells was used for Affymetrix GeneChip analysis (Affymetrix, Santa Clara, CA). The integrity of the mRNA was checked by agarose gel electrophoresis, 200 ng of mRNA was processed, and 15 ug of cRNA was hybridized to Affymetrix mouse MOE430A array chips according to the manufacturer’s recommended protocol.
Gene expression profiles were compared between the GnRH-treated R/P and untreated R/P groups. Three independent experiments were analyzed, resulting in a total of twelve microarray sets (six R/P sets). Affymetrix chip image files were converted to probe set data (.CEL files) using the Gene Chip Operating System (GCOS). The MOE430A Affymetrix chip consists of 22,690 probes. 6556 probes had Absent Calls that were consistent across all twelve chips and were removed, leaving 16,134 probes for analysis. Raw expression values in the CEL files were pre-processed via the Robust Multichip Average (RMA) method (Irizarry et al., 2003), as implemented in Bioconductor (www.bioconductor.org), a suite of programs for the R statistical programming language (http://cran.r-project.org). RMA default values were employed to pre-process the twelve chips simultaneously.
The log2 gene expression values obtained from RMA pre-processing were analyzed as follows. First, genes that were differentially expressed between the GnRH-treated R/P and untreated R/P groups were identified via two different criteria: fold change and t-statistics. Fold-change analysis highlights potentially biologically interesting genes. The t-statistics were used to compute the magnitude of the variation among biological replicates as compared to the variation between the treated and untreated groups. A large absolute value in the t-statistic is indicative that there is larger statistical variation between treated and untreated groups than variation within a specific group. A composite measure of fold-change and t-statistics, called bioweight (Rosenfeld et al., 2004), was used to produce a list of the top 5% (800) of the most regulated mRNA species amongst those that are found to be expressed in LβT2. The bioweight measure is a product of the across-replicate fold change and the negative log10 of p-values for the gene specific t-tests. The advantage of bioweight analysis is that the impacts of fold-change analysis and gene specific t-test analysis are studied simultaneously.
Confirmatory differential gene expression analysis was conducted via the permutation method Significance Analysis of Microarrays (SAM) (Tusher et al., 2001). The SAM method detects mRNA species with statistically significant differences in expression by computing a score for each species relative to the species-specific standard deviation (Tusher et al., 2001). Individual mRNAs with scores greater than an adjustable threshold are declared significant. The percentage of mRNAs that are identified by chance is the False Discovery Rate (FDR), which was set at 0.2. Since non-parametric, permutation-based techniques, such as SAM, are hindered by experiments with small sample sizes, it is reasonable to compare the results from SAM to other methods such as bioweight. The SAM technique was computed in Bioconductor via the siggenes library. The q-values generated by SAM are an FDR-adjusted equivalent to the p-value (Storey, 2002).
Confirmatory analysis employed unsupervised clustering to determine if the treatment groups could be recovered. Dendrograms of expression patterns and samples were computed via hierarchical clustering using complete-linkage as the between-cluster distance criteria and Pearson’s correlation as the similarity measure (Eisen et al., 1998). Heatmaps (false color images) of the top 800 and bottom 800 bioweight mRNAs were generated such that the rows (samples) and columns (genes) were clustered independently using hierarchical clustering via Pearson’s correlation for within-cluster distance and complete linkage for between-cluster distance. Red indicates strong positive correlation and green indicates strong negative correlation (Fig. S2).
The top regulated mRNAs were further characterized by the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.kegg.org) via the Bioconductor package, annffy (Gentleman et al., 2005). The annffy package is designed to interface between Affymetrix chip analysis results and web-based databases by gathering annotation data from diverse resources. Probes that were not associated with a KEGG pathway (approximately 400) were not summarized in Figure S1. mRNAs with multiple probes or that were associated with multiple pathways were consolidated.
2.7. RNA-binding Protein Immunoprecipitation
RNA-binding protein immunoprecipitation (RIP) was performed to analyze the interaction of DUSP1 mRNA with HuR as described previously (Jain et al., 2011). In brief, LβT2 cells were treated with vehicle or 10 nM GnRH for 30 min and then lysate with PEB containing 1% Triton-X. After centrifugation, the supernatants were pre-cleared with 5 μg of normal mouse IgG (sc-2025, Santa Cruz Biotechnology, Santa Cruz, CA) and 50 μl of protein G Dynabeads (Invitrogen, Carlsbad, CA) for 40 min at 4° C with constant mixing. After remove the beads, 10 μl supernatant was used to as internal controls of Input RNA. For coated magnetic bead with antibody, 50 μl of protein G Dynabeads (Invitrogen, Carlsbad, CA) were washed with 1 ml of 0.1 M citrate-phosphate buffer (pH 5.0) and mixed with 5 μg of mouse anti-HuR antibody (sc-5261, Santa Cruz Biotechnology, Santa Cruz, CA), or 5 μg of mouse IgG (sc-2025, Santa Cruz Biotechnology, Santa Cruz, CA) for 30 min at room temperature with constant mixing. The HuR- and mouse IgG antibody-coated beads were then resuspended in 400 μl of the NT2 buffer (50 mM Tris (pH 7.4), 1 mM MgCl2, 150 mM NaCl, 0.05% Nonidet P-40) supplemented with 100 U/ml RNase OUT, 1 mM DTT, and 20 mM EDTA, and then incubated for 3 h at 4° C with 100 μl of the cell lysate. After beads were washed 5 times using NT2 buffer, and then incubated for 30 min at 55 °C in 150 μl of NT2 buffer supplemented with 1.2 mg/ml of proteinase K and 1% % SDS. The immunoprecipitated RNA was extracted using phenol/chloroform and analyzed with reverse transcription-PCR (RT-PCR) using above DUSP1 primers.
2.8. Statistical Analysis
The areas under the curves of the polyribosomal profiles were integrated using SigmaPlot v9.01 (Systat Software, Richmond, CA). To correct for the skewed distribution of ratio data, all ratio data was transformed using log2 and a value of 10 was added to ensure positive values prior to analysis. All subsequent statistical analysis was conducted using JMP (SAS Institute, Cary, NC) on untransformed (other than log2 + 10 for ratio or fold-redistribution values) or optimally Box-Cox transformed values. Redistribution data is analyzed as fold-change ratio. Data obtained from transfection experiments was evaluated by multifactor ANOVA and post hoc testing with Tukey’s multiple comparison test. Except for the microarray data set, all the experiments were repeated at least four independent times and reported values are represented by the means +/− SEM of those experiments.
2. Results
3.1. Differential sensitivity to translation inhibition
To understand translational regulation by GnRH stimulation, we examined the global redistribution of mRNA between actively translating polyribosome and non-translating RNP complexes in LβT2 gonadotropes by subcellular fractionation on sucrose gradients after treatment with GnRH or vehicle for 30 minutes. Our previous observations showed that mRNA displacement was transient with a maximal effect at 30 minutes after a 5- minute exposure to GnRH that resolved within 60 minutes. After isolating mRNA from pooled fractions encompassing either the polyribosome or RNP pools (Fig. 1A), we examined the relative change in abundance of individual mRNAs in the polyribosome and RNP pools by microarray. Data were processed as described in the Materials and Methods and the change of individual mRNA species in each pool after GnRH treatment was expressed as fold change relative to vehicle treatment.
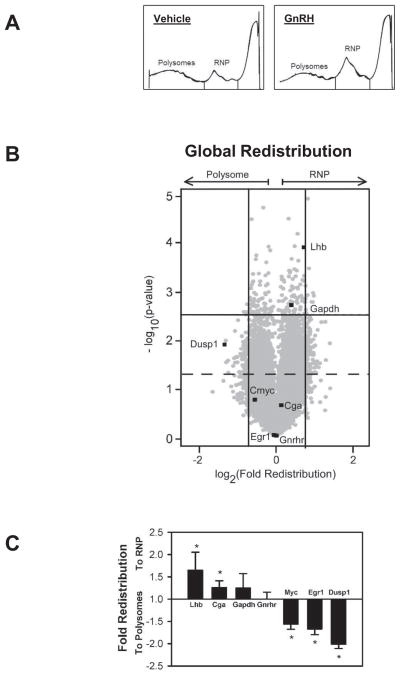
Figure 1. GnRH causes a targeted redistribution of mRNA in polyribosome and ribonucleoprotein complexes in LβT2 gonadotrope cells.
A) a sample profile of fractionated cell extracts from LβT2 cells treated with vehicle or GnRH at 10 nM for 30 minutes. Extracts were fractionated on 10–50% sucrose gradients and collected from the bottom to the top (50%–10%, left to right) while absorbance was monitored at 254 nM wavelength. Area under the curve containing polyribosomes and single ribosome/RNP fractions are indicated by the vertical lines through the chart. These areas were collected and purified mRNA derived from these fractions were designated as polyribosome or RNP fractions for subsequent studies. B) Bioweight plot of changes in mRNA representation in polyribosome and RNP fractions of LβT2 cells. The method of analysis is described in Materials and Methods. Briefly, mRNA isolated from polyribosome and RNP fractions from three independent experiments was subject to microarray quantitation and analysis. Change in mRNA representation between pools is plotted by Log2 fold change on the abscissa and −log10 of the Student’s test p-value on the ordinal axes to illustrate overall shifts in representation. Individual mRNAs examined in this study are highlighted and designated in the plot. Gray dots represented individual mRNA probes. Vertical bars represent 95% confidence interval for the fold redistribution, Horizontal bar represents 95% cutoff for −Log10 p-value. Approximately 800 mRNAs fall outside these regions. The dashed line indicates significant change from control at a=0.05. 1362 probes showed significant change from vehicle treated control values. C) Results of quantitative PCR measuring fold redistribution of individual mRNAs after GnRH treatment identified in the microarray analysis above. Bars above the line represent fold change to the RNP fraction and bars below the line represent fold change to the polyribosome (polysomes) fraction from three (Dusp1) or six (Lhb, Cga, Gapdh, Gnrhr, Myc, Egr1) replicates. Asterisks indicate significant change from untreated control levels by Student’s t-test.
We examined the effect of mRNA distribution between the polyribosome and RNP pools using bioweight measurements of individual species (Rosenfeld et al., 2004). In this method, individual mRNAs are ranked by relative change from control and by significance of the measured change to distinguish the mRNAs most influenced by the treatment. At the global level, RNP accumulation was evident after GnRH treatment, consistent with our previous observation that GnRH causes a transient attenuation through activation of the UPR. However, by examining mRNAs that are redistributed, it is evident that invocation of the UPR by GnRH does not result in a generalized loss of mRNA from the actively translating pool of polyribosomes. As indicated by the plot in Figure 1B, all mRNAs significantly changed by GnRH treatment are indicated by those appearing above the dashed line in figure 1B. This represents 1362 distinct probes corresponding to unique mRNA species, including variants of the same gene. Distinct populations of mRNAs are enriched in either the polysome or RNP pools by GnRH treatment, as indicated by separation on the abscissa from zero, which indicates no change. This is particularly evident in the clustering analysis and heat map representation of the top 800 mRNAs (representing 774 genes) affected by GnRH treatment displayed in Figure S1A. Increased representation in the RNP pool indicates a relative decrease in association with ribosomes and reduced utilization by the translational machinery. Increased relative abundance in the polysome pool indicates an increase in association with ribosomes, suggesting increased utilization by the translational machinery. This analysis confirmed our previous observations that Lhb mRNA is displaced to the RNP pool and this analysis shows it is one of the most highly affected mRNAs (Table 1, Supplemental Table 1). Previous analysis has shown that increased representation in the RNP pool of Lhb, Cga, and Gapdh mRNA is not a result of increased mRNA content due to increased transcription (Do et al., 2009), but this has not been generally confirmed for all affected mRNAs.
Table 1. Top 10 genes by bioweight enriched in polyribosome or RNP fractions by GnRH.
Table 1. Top 10 genes enriched in the Ribonucleoprotein (RNP) and polyribosome cytosolic fraction of cells as determined by microarray analysis and bioweight method of fold change from control. The Bioweight index is determined by the product of absolute average fold change from control and negative logarithm of the t-statistic p-value. The top genes listed below are abstracted from a full list of the top 800 genes (top 5% of those measured) presented as Table S1 in supplementary data..
| Top 10 mRNAs enriched in the RNP fraction by GnRH
| ||||
|---|---|---|---|---|
| Bioweight | Gene | Common Name | Accession | Probe |
| 4.07 | Ndufa3 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 | AV048277 | 1452790_x_at |
| 3.04 | Ecel1 | endothelin converting enzyme-like 1 | NM_021306 | 1422586_at |
| 2.98 | 2900010M23Rik | RIKEN cDNA 2900010M23 gene | NM_026063 | 1448685_at |
| 2.86 | Mknk2 | MAP kinase-interacting serine/threonine kinase 2 | NM_021462 | 1418300_a_at |
| 2.86 | Mrps15 | mitochondrial ribosomal protein S15 | BB314055 | 1456109_a_at |
| 2.84 | Lhb | luteinizing hormone beta | NM_008497 | 1450795_at |
| 2.79 | Aplp1 | amyloid beta (A4) precursor-like protein 1 | NM_007467 | 1416134_at |
| 2.70 | Srf | serum response factor | BI662291 | 1418255_s_at |
| 2.66 | Atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | AV066932 | 1437164_x_at |
| 2.58 | Ndufa3 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 | AV048277 | 1428464_at |
| Top 10 mRNAs enriched in the Polyribosome fraction by GnRH
| ||||
|---|---|---|---|---|
| Bioweight | Gene | Common Name | Accession | Probe |
| 2.58 | Dusp1 | dual specificity phosphatase 1 | NM_013642 | 1437354_at |
| 2.54 | 9630042H07Rik | RIKEN cDNA 9630042H07 gene | BC027796 | 1415823_at |
| 2.36 | Catnb | catenin beta | NM_007614 | 1424766_at |
| 2.32 | N/A | 0 day neonate cerebellum cDNA, RIKEN, clone:C230091D08 | AI154956 | 1429144_at |
| 2.14 | Scd2 | stearoyl-Coenzyme A desaturase 2 | BG060909 | 1419277_at |
| 2.04 | BC004701 | cDNA sequence BC004701 | BC004701 | 1419529_at |
| 1.99 | 2310032D16Rik | RIKEN cDNA 2310032D16 gene | AV291259 | 1418025_at |
| 1.98 | Usp31 | ubiquitin specific protease 31 | BG068704 | 1415996_at |
| 1.98 | Il23a | interleukin 23, alpha subunit p19 | NM_031252 | 1420922_at |
| 1.97 | Bhlhb2 | basic helix-loop-helix domain containing, class B2 | NM_011498 | 1434392_at |
In contrast, many mRNAs are enriched in the polysome pool after GnRH treatment, indicating increased association with ribosomes and translational utilization. Included in this population is Dusp1 mRNA. DUSP1 participates in the dephosphorylation of MAP kinases after GnRH stimulation and we have shown that protein levels increase after GnRH stimulation, consistent with this redistribution (Nguyen et al., 2010). It is also noted that c-myc was also increased in abundance in the polysome fraction. Unlike Dusp1 mRNA, translation of C-myc mRNA occurs via a cap-independent translational mechanism involving internal ribosome entry (Nanbru et al., 1997). The presence of mRNAs utilizing both cap-dependent and cap-independent translation initiation mechanisms suggests that enrichment of mRNA in the polyribosome pool occurs through a mechanism that is not determined by the mode of ribosomal assembly and translation initiation. The relative distribution of Egr1 mRNA between pools is not reliably determined by microarray analysis. We and others have documented a rapid increase in total mRNA abundance after GnRH treatment (Lawson et al., 2007; Ruf and Sealfon, 2004), initial levels are low and affect the reproducibility of the assessment. This is more closely evaluated below.
A more generalized view of the consequences of mRNA redistribution can be obtained by examination of the cellular pathways affected. Evaluation by KEGG pathways of the top 800 mRNAs subject to redistribution is shown in supplemental Figure S1B. Of the 800 top regulated mRNAs, only about half had KEGG pathway information available. The enrichment of mRNAs in the RNP pool identifies those that are potential targets of translational inhibition by induction of the UPR and subsequent phosphorylation of EIF2A, which blocks initiation of assembled translational complexes. Examination of RNP-displaced genes revealed a number of important features. The abundance of genes associated with subcellular structures or membranes is evident in the proportion that is displaced to the RNP fraction by GnRH. These include those encoding proteins in the oxidative phosphorylation pathway, gap junction, tight junction and adherens junction pathways as well as protein export and ribosomal proteins themselves. The mRNAs subject to accumulation in the RNP may also be a consequence of the distinct subcellular ER and mitochondrial localization of EIF2AK3 after induction of the UPR (Kondratyev et al., 2007; Verfaillie et al., 2012).
The analysis also revealed pathways known to be important for GnRH signaling, including MAPK, and insulin. For MAPK, mRNAs enriched in the RNP pool were immediate signal transducers (Map4k4, Mapkapk2, Mknk2, Ras), indicating a pause in synthesis of these factors, while feedback regulators (Dusp1, Pla2g2d) were enriched in the polysome pool, indicating either a resistance to UPR control mechanisms or specific enrichment via other regulatory processes. The opposite effect was observed for members of the JAK-STAT signaling pathway. Viewed in the context of signal induction and feedback control, individual components of signaling cascades may be regulated at the level of translation by either attenuation through induction of the UPR or increased synthesis through enrichment in the polyribosome pool.
3.2. Enrichment of specific mRNAs to the polyribosome and RNP pools
To validate these results we examined the redistribution of specific mRNA species central to the regulation of gonadotropes by GnRH. We monitored Lhb, Cga, Gapdh, Gnrhr, Egr1, and Dusp1 representation in ribosome or RNP complexes after GnRH treatment using quantitative real time PCR. Consistent with the microarray analysis above and previous observations, the relative increase in Lhb, Cga, and Gapdh in the RNP fractions was confirmed (Fig. 1C). Evaluation of Gnrhr mRNA indicated no significant change from GnRH treatment. A GnRH-induced increase in the relative abundance of Dusp1 and Egr1 mRNA in the polyribosome pool was also confirmed. Additionally we examined C-myc mRNA, which is translated via internal ribosome entry rather than the 5′ cap-dependent mechanism. After GnRH treatment C-myc was also increased in the polyribosome fraction. Because translation of C-myc is not cap-dependent, increased presence in the polyribosomal fraction suggests that enrichment occurs through a mechanism other than cap-dependent translation initiation. Thus three populations of mRNA are identified in this analysis; one showing increased relative representation in polyribosome complexes after stimulation with GnRH indicating consistent or increased translational utilization, one largely unaffected, and a third showing a relative increase in the RNP fractions, indicating decreased translational utilization.
3.3. Activation of the unfolded protein response does not explain Duspl mRNA redistribution
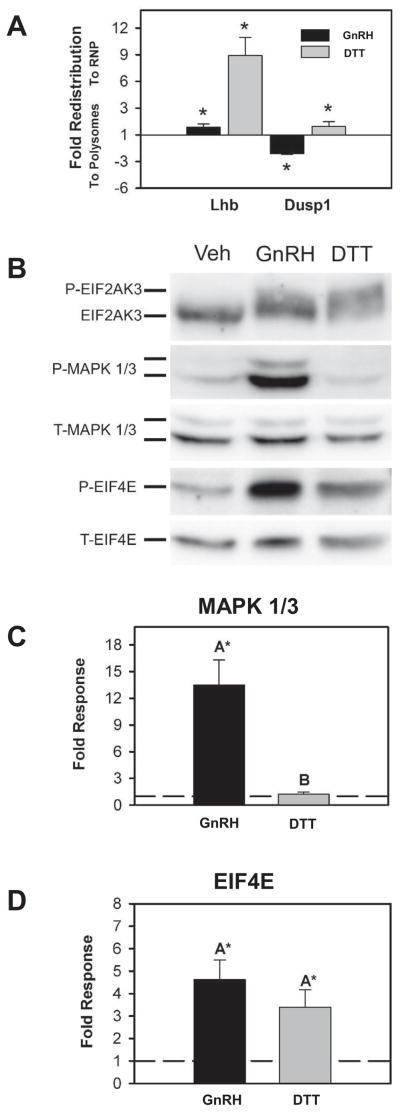
GnRH receptor occupancy leads to stimulation of a number of downstream signaling factors that participate in the initiation of secretion, increased transcriptional activity, increased translation, and modulation of ER function through the UPR. Translational attenuation by the UPR occurs due to phosphorylation of EIF2A by EIF2AK3. However, as demonstrated above, not all mRNAs appear to be susceptible to inhibition. In particular Dusp1 mRNA appears to be resistant to attenuation. To determine whether Dusp1 enrichment in the polyribosome fraction was a result of UPR activation, we compared the effect of GnRH on Dusp1 mRNA to the direct chemical activation of the UPR by DTT, a known activator of the UPR. DTT treatment results in a near complete inhibition of translation. Comparison of GnRH and DTT effects on Lhb and Dusp1 mRNA showed that under strong UPR activation by DTT, Lhb and Dusp1 mRNA are both enriched in the RNP fraction (Fig. 2A). However, GnRH treatment only affects Lhb mRNA. Examination of EIF2AK3 phosphorylation in both DTT and GnRH treated LβT2 cells by Western blot shows that both treatments resulted in EIF2AK3 activation (Fig. 2B). In contrast GnRH caused a significant increase in MAPK1/3 phosphorylation whereas DTT had no effect. Phosphorylation of the translation initiation factor EIF4E was also increased by both DTT and GnRH, indicating some independence from MAPK 1/3 activation (Fig. 2 B&C). Therefore the increased abundance of Dusp1 mRNA in the polyribosome fraction cannot be explained by activation of the UPR or of EIF4E and suggests that other components of the GnRH signaling response are responsible.
Figure 2. Activation of the UPR does not explain polyribosome enrichment of Dusp1 mRNA.
A) Summary of Lhb and Dusp1 enrichment in RNP and polyribosome fractions of cytoplasmic extracts from vehicle or 10nM GnRH-treated LβT2 cells with or without pretreatment with 2 mM DTT. DTT causes enrichment of both Lhb and Dusp1 mRNA to the RNP fraction, but GnRH causes RNP enrichment of Lhb mRNA and polyribosomal enrichment of Dusp1 mRNA. Asterisks indicate significant fold change from untreated samples in four independent experiments as determined by Student’s t-test. B) An example Western blot analysis of signaling proteins isolated from Vehicle (Veh), GnRH and DTT treated LβT2 cells using antibodies directed against EIF2AK3, phosphorylated MAPK1/3, total MAPK 1/3, phosphorylated EIF4E, and total EIF4E. Proteins were visualized by chemiluminescent imaging and resulting images used in quantification of individual proteins as below. Images are presented in inverted grayscale. C) Summary of Western blot quantification of phosphor- and total MAPK1/3 from 4 replicate experiments showing increased phospho-MAPK1/3 to total MAPK 1/3 ratio due to GnRH, but not DTT treatment of LβT2 cells. Protein levels were determined by chemiluminescent imaging and quantification. Phosphorylated to total ratios of protein were normalized to untreated levels and plotted. GnRH, but not DTT, increases MAPK1/3 phosphorylation. D) Summary of EIF4E phosphorylation in response to GnRH and DTT treatment of LβT2 cells as above. Ratios of phospho- to total- EIF4E were determined and normalized to untreated controls in five independent experiments. Both GnRH and DTT increase EIF4E phosphorylation to a similar degree. In both C and D dashed lines represent control ratio of phospho-MAPK1/3 to total MAPK 1/3 or phospho-EIF4E to total-EIF4E normalized to one. Asterisks indicate significant difference from untreated control as determined by Student’s t test and groups with different letters are significantly different from each other as determined by two-sample t test.
3.4. Dusp1 mRNA redistribution is recapitulated by activation of PKC
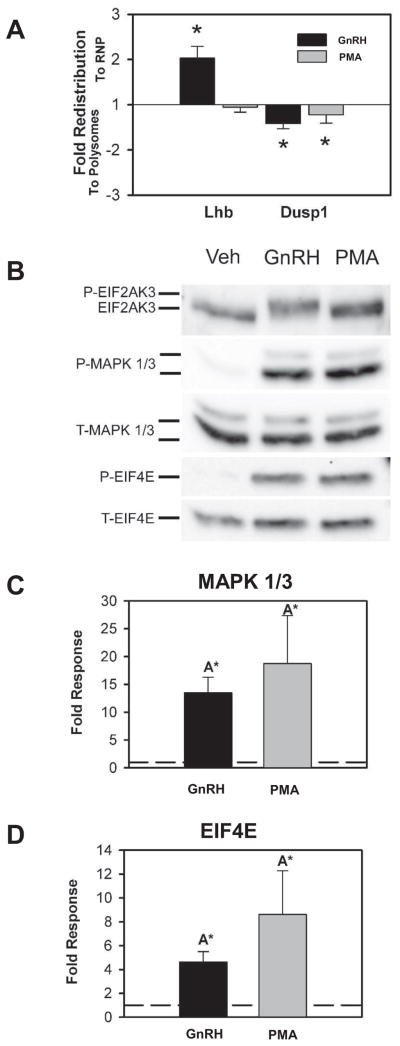
Dusp1 enrichment in polyribosomes did not appear to be dependent on UPR activation or phosphorylation of EIF4E, suggesting that signaling cascades other than those targeting translational control by the UPR or cap-dependent translation initiation via EIF4E activation may contribute to the behavior of Dusp1 mRNA. GnRH, but not DTT treatment, strongly activates MAPK 1/3 and this correlated with polyribosome enrichment of Dusp1 mRNA. This suggests that activation of the PKC-MAPK signaling pathway by GnRH may contribute to the observed behavior of Dusp1 mRNA. Activation of protein kinase C isoforms by PMA strongly activates MAP kinases including MAPK1/3, and this recapitulates PKC activation by GnRH and regulation of gonadotropin subunit genes (Dobkin-Bekman et al., 2010; Vasilyev et al., 2002). Treatment of LβT2 cells with PMA resulted in enrichment of Dusp1 mRNA in the polyribosome fraction but did not significantly affect Lhb mRNA distribution relative to vehicle-treated cells (Fig. 3A). We examined the effect of PMA on EIF2AK3 activation by Western blot and found that PMA had no measureable effect on phosphorylation (Fig. 3B). However, PMA treatment did significantly increase both MAPK1/3 and EIF4E phosphorylation (Fig 3B&C). Because EIF4E is also phosphorylated in response to UPR activation by DTT and this does not lead to Dusp1 mRNA enrichment in polyribosomes, these observations combined with those in Figure 2 support the interpretation that translational control mechanisms involving MAPK1/3 activation contribute to the polyribosomal enrichment of Dusp1 by GnRH.
Figure 3. Polyribosome enrichment of Dusp1 mRNA is recapitulated by treatment with PMA.
A) Summary of redistribution of Lhb and Dusp1 mRNA to RNP and polyribosome fractions in response to GnRH and PMA treatment. Both GnRH and PMA induce polyribosomal redistribution of Dusp1 mRNA, but PMA has no effect of Lhb distribution. Asterisks indicate significant change from control from three independent experiments as determined by Student’s t test. B) Example Western blot images from LβT2 cells treated with Vehicle (Veh) GnRH, or PMA showing a lack of EIF2AK3 phosphorylation but increased MAPK1/3 and EIF4E phosphorylation. Chemiluminescent images are displayed in inverted grayscale. C) Summary of three independent Western blot analyses of MAPK 1/3 phosphorylation in GnRH and PMA treated cells. Both GnRH and PMA increase MAPK 1/3 phosphorylation to a similar degree. D) Summary of three independent Western blot analyses of EIF4E phosphorylation in response to GnRH and PMA treatment. GnRH and PMA both increase EIF4E phosphorylation to a similar degree. In both C and D dashed lines represent control ratio of phospho-MAPK1/3 to total MAPK 1/3 or phospho-EIF4E to total-EIF4E normalized to one. Asterisks indicate significant difference from untreated control as determined by Student’s t test and groups with different letters are significantly different from each other as determined by two-sample t test.
3.5. Dusp1 mRNA redistribution is sensitive to MAPK1/3 inhibition
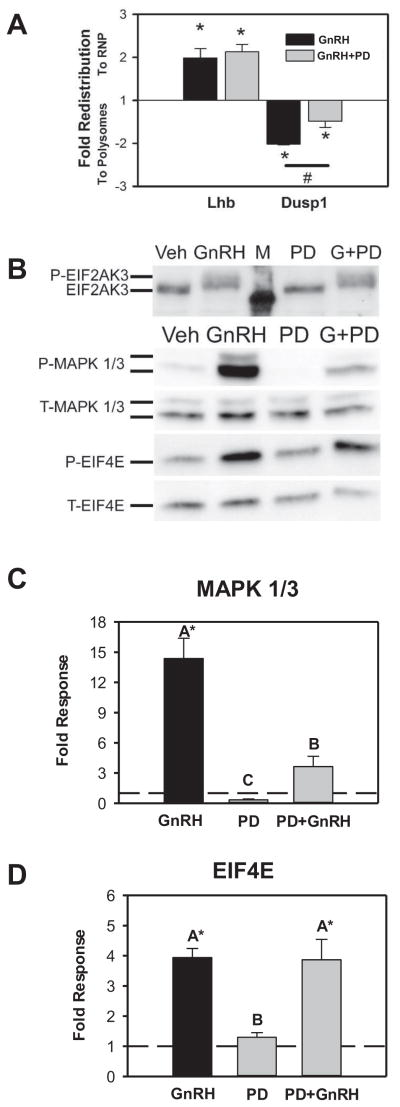
Both GnRH and PMA strongly activate MAPK1/3 and this corresponds with their ability to induce Dusp1 mRNA redistribution. Therefore we tested the dependence of Dusp1 redistribution on MAPK1/3 activation. We treated LβT2 cells with the MEKK inhibitor PD098059 to block GnRH activation of MAPK1/3 prior to GnRH treatment and fractionation on sucrose gradients. Treatment with PD098059 resulted in attenuation of Dusp1 mRNA enrichment in the polyribosome fractions after GnRH treatment but has no effect on Lhb mRNA enrichment in the RNP fraction, indicating that polyribosome enrichment occurs through a MAP kinase-dependent mechanism that is independent of the mechanisms contributing to enrichment of Lhb mRNA in the RNP fractions (Fig. 4A). PD098059 does not impact EIF2AK3 or EIF4E activation by GnRH (Fig. 4B). However, MAPK1/3 activation was significantly attenuated by pretreatment. This supports the role of MAP kinase signaling in polyribosome enrichment of Dusp1 mRNA.
Figure 4. Polyribosome enrichment of Dusp1 mRNA is attenuated by inhibition of the MAPK1/3 signaling cascade.
A) Summary of redistribution of Lhb and Dusp1 mRNA pretreated with vehicle or with 30μM PD098059 for 30 minutes prior to a 30 minute stimulation with vehicle or 10nM GnRH. A summary of the independent determinations of fold change relative to vehicle-treated controls is plotted. Asterisks indicate significant change from control from three independent experiments as determined by Student’s t test. B) Example Western blot images from LβT2 cells treated with vehicle (Veh), GnRH, PD 098059 (PD), or GnRH+PD098059 (G+PD). M indicates molecular weight marker. Images show activation of EIF2AK3 and EIF4E by GnRH is not affected by PD098059 pretreatment but MAPK 1/3 phosphorylation is attenuated. Chemiluminescent images are displayed in inverted grayscale. C) Summary of three independent Western blot analyses of MAPK 1/3 phosphorylation in GnRH, PD098059 or PD098059 plus GnRH-treated cells. GnRH-induced phosphorylation of MAPK1/3 is attenuated by PD098059. D) Summary of three independent determinations of GnRH-induced phosphorylation of EIF4E either alone or in the presence of PD098059 showing no effect on phosphorylation. In both C and D dashed lines represent control ratio of phospho-MAPK1/3 to total MAPK 1/3 or phospho-EIF4E to total-EIF4E normalized to one. Asterisks indicate significant difference from untreated control as determined by Student’s t test and groups with different letters are significantly different from each other as determined by ANOVA and post hoc analysis with Tukey’s HSD test.
3.6. Dusp1 mRNA redistribution is insensitive to rapamycin
The above results indicate that Dusp1 mRNA enrichment in the polyribosome fractions does not require activation of cap-dependent initiation. EIF4E phosphorylation depends on its release from an inactive complex with 4E binding protein (4EBP1). Phosphorylation of 4EBP1 by the growth regulatory kinase mTOR releases EIF4E, enabling interaction with mRNA cap structures and phosphorylation by the translational initiation machinery. To examine the dependence of Dusp1 enrichment in polyribosomes on the cap-dependent translation initiation mechanism directly, we examined the effect of the mTOR inhibitor rapamycin on GnRH-induced mRNA redistribution. Inhibition of with mTOR with rapamycin inhibits phosphorylation of 4EBP1 thus blocking cap-dependent translation initiation. The presence of rapamycin reduced the overall content of mRNA in the polyribosome pool (Fig. 5A). However, GnRH treatment still caused an enrichment of Dusp1 in that pool and rapamycin had no effect on Lhb enrichment in the RNP pool. Although rapamycin inhibited phosphorylation of 4EBP1 by GnRH, there was no effect on GnRH induced phosphorylation of MAPK 1/3 (Fig. 5B, 5C). As we and others have reported, rapamycin does not completely inhibit EIF4E phosphorylation in response to a signaling stimulus (Beretta et al., 1996; Nguyen et al., 2004). Overall this result confirms that activation of EIF4E and cap-dependent translation does not contribute to increased DUSP1 representation in the polyribosome pool.
Figure 5. Polyribosome or RNP enrichment of Lhb or Dusp1 mRNA by GnRH is not affected by rapamycin.
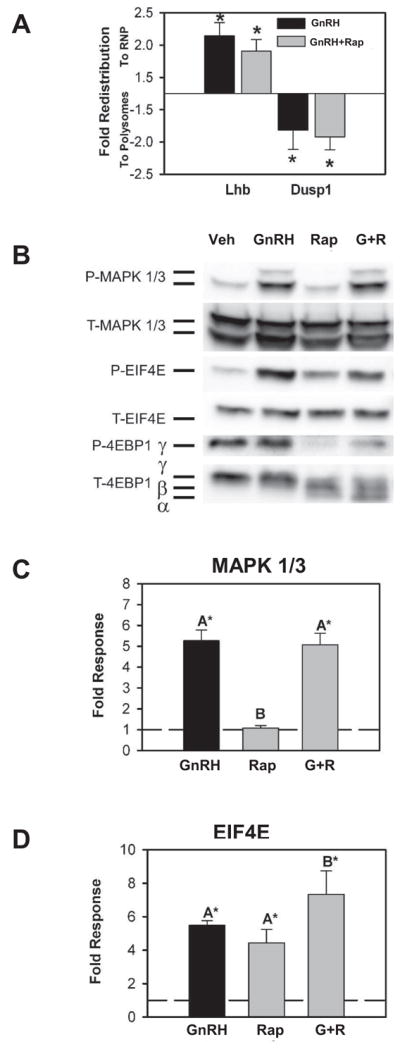
A) Summary of three independent determinations of polyribosome or RNP enrichment of Lhb and Dusp1 mRNA by GnRH either alone or with 30 minute pretreatment with the mTOR inhibitor rapamycin showing that neither Lhb nor Dusp1 enrichment is sensitive to inhibition of cap-dependent translation. Phospho-4EBP is reduced by pretreatment with rapamycin and the hypo- and un-phosphorylated α, β and γ isoforms are increased in abundance. Chemiluminescent images are presented in inverted grayscale. C) Summary of three independent Western blot determinations of MAPK 1/3 phosphorylation by GnRH after pretreatment with vehicle or rapamycin showing no effect on basal or GnRH-induced levels of phospho-MAPK 1/3. D) Summary of three independent determination of EIF4E phosphorylation after pretreatment with vehicle or rapamycin showing a modest increase in EIF4E phosphorylation in response to GnRH in both vehicle and rapamycin-treated cells. In both C and D dashed lines represent control ratio of phospho-MAPK1/3 to total MAPK 1/3 or phospho-EIF4E to total-EIF4E normalized to one. Asterisks indicate significant difference from untreated control as determined by Student’s t test and groups with different letters are significantly different from each other as determined by ANOVA and post hoc analysis with Tukey’s HSD test.
3.7. The Dusp1 3′UTR confers resistance to translational inhibition
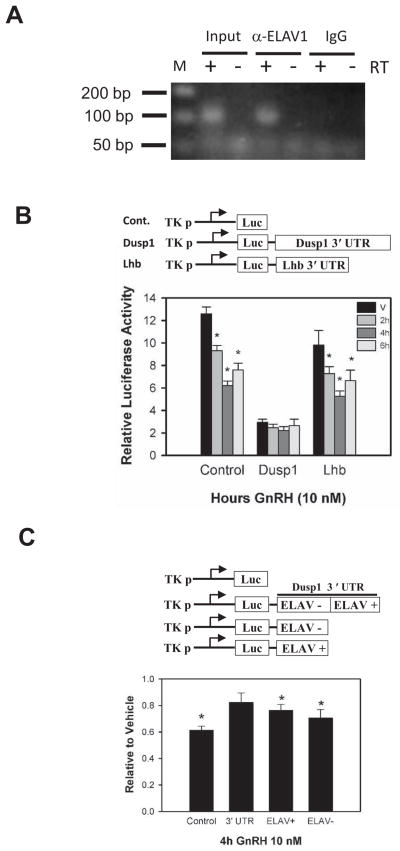
To understand which mechanisms may be responsible for enrichment of Dusp1 mRNA in polyribosomes, we explored the structure of the mRNA. The Dusp1 mRNA is predicted to be a structurally complex molecule that interacts with a number of RNA-binding proteins including tristetraproline/ZFP36 and ELAV1/HuR which bind an AU-rich element in the distal 3′UTR (Emmons et al., 2008; Kuwano et al., 2008; Lin et al., 2008). We confirmed the interaction of ELAV1 with Dusp1 mRNA by RNA-IP using an antibody directed against mouse ELAV1 followed by RT-PCR with Dusp1-specific primers (Fig. 6A).
Figure 6. The 3′ UTR of Dusp1 mRNA is bound by ELAV1 and confers resistance to GnRH-induced reduction of expression in LβT2 gonadotrope cells.
A) Immunoprecipitation of LβT2 cell extracts with anti ELAV1 antibody (α-ELAV1) followed by RT-PCR using primers directed against Dusp1 mRNA shows interaction between ELAV1 and Dusp1 mRNA. Input, direct RT-PCR of LβT2 cell extracts. IgG, immunoprecipitation with normal mouse IgG. +/− indicates presence or absence of reverse transcriptase in the cDNA reaction prior to PCR. M designates marker lane B) Structure of reporter genes bearing the encoded pGL3 SV40 poly-Adenylation site without an encoded 3′UTR (Cont., Control), the 3′UTR from Dusp1 (Dusp1), or the 3′UTR from Lhb (Lhb). Reporter plasmids were co-transfected into LβT2 cells with an internal control plasmid expressing β-galactosidase. Cells were stimulated with 10nM GnRH for the times indicated and assayed for luciferase and β-galactosidase activity. The ratio of Luciferase to β-galactosidase is plotted in the figure. Asterisks indicate significant difference from the time vehicle control from four independent experiments as determined by ANOVA and Tukeys’ HSD post hoc test. C) Reporter plasmids bearing no 3′ UTR (Control), the Dusp1 3′UTR (3′UTR), the Dusp1 3′UTR with a deletion of the region containing the predicted sequence and secondary structure interacting with ELAV1 (ELAV −) or the remaining region of the Dusp1 3′UTR were co-transfected with a control vector expressing β-galactosidase into LβT2 cells and treated with GnRH for 4h. Cells were harvested and assayed for luciferase and β-galactosidase activity and plotted in the chart. Asterisks indicate significant difference from vehicle-treated control level normalized to one as determined by Student’s t-test of four independent experiments.
To evaluate the effect of the Dusp1 3′UTR on gene expression, we constructed three reporter genes of identical plasmid backbone, the 80bp proximal TK promoter, and the firefly luciferase gene, that differ only in the composition of the 3′ UTR. One contained only the reporter sequences up to the SV40 polyadenylation signal, the second included the Lhb 3′UTR and the third contained the full Dusp1 3′ UTR. Each of these was individually transfected into LβT2 cells with an internal control plasmid containing the same plasmid backbone but including the CMV immediate-early promoter and the β-galactosidase reporter gene. Earlier studies showed that GnRH-induced translational pausing is transient (Do et al., 2009). Redistribution peaks at 30 minutes after GnRH stimulation and resolves at 60 minutes after stimulation. If enrichment of mRNA into the RNP pool is due to decreased translation, a transient loss of translational activity would be accompanied by a corresponding decrease in reporter gene activity after GnRH treatment. To examine the possibility that the Dusp1 3′ UTR confers resistance to translational pausing, transfected cells were treated with GnRH and harvested at 2h, 4h, and 6h after treatment. The half-life of firefly luciferase in eukaryotic cells has been measured at approximately 3.7 h (Leclerc et al., 2000). Consistent with the measured half-life of luciferase and the occurrence of maximal translational pausing at 30 minutes after GnRH stimulation, we observed a significant decrease in luciferase activity derived from the control and Lhb 3′UTR reporter genes 4h after GnRH stimulation (Fig. 6B). In contrast, the reporter construct bearing the Dusp1 3′UTR was resistant to the GnRH-induced decrease in luciferase activity, although relative activity of the reporter gene to the internal control was lower. To test whether the ELAV1 binding region of the Dusp1 mRNA alone was sufficient to confer resistance to GnRH-induced inhibition of translation, we examined deletion constructs of the 3′ UTR. Although the intact 3′UTR is able to confer resistance to translational inhibition by GnRH, the ELAV1 binding region alone or the 3′UTR minus the ELAV1 binding region were both sensitive to GnRH-induced repression of reporter gene expression (Fig. 6C). Thus the entire 3′UTR may be necessary to confer resistance, or deletion of portions of the 3′UTR are disruptive to the overall structure of the 3′UTR and prevent proper function.
4. Discussion
The regulation of translation allows for rapid, reversible, and fine control of gene expression in response to changes in the cellular environment. This regulation may involve either changes that affect mRNAs at the global level, or those that affect a subset of mRNAs in response to changes in environment or to extracellular signals. (Dang Do et al., 2009; Kuhn et al., 2001; Mathews et al., 2000). Subcellular localization of translation is also a component of regulation, as in the case of distal axonal translation of numerous mRNAs (Deglincerti and Jaffrey, 2012), and regulation of the UPR translational response is also localized to specific areas associated with the ER and mitochondria (Verfaillie et al., 2012). Translational control can operate selectively through dependence on the cap-binding translation initiation complex or through independence from cap-dependent translational initiation by utilization of internal ribosomal entry for initiation. Additional mechanisms contributing to the utilization of mRNA by the translational machinery include sequestration in stress granules or processing bodies, interaction with 3′ UTR binding proteins and targeting by miRNAs. All of these processes can be affected by extracellular signals or changes in physiological status.
Altogether these regulatory mechanisms comprise a complex network that can operate independently of transcriptional regulation, providing a layer of gene regulation that may not be evident by direct examination of transcriptional regulation or examination of steady-state mRNA levels in cells. It has become clear that observed transcriptional changes do not strictly correspond in timing or magnitude to observed changes in protein levels demonstrated in both in vivo and in vitro studies. One striking example of this includes the modest and delayed increase in Lhb mRNA in response to pulsatile GnRH in rats. Measurements in vivo show only moderate increases in Lhb mRNA levels (Burger et al., 2002), although primary transcript levels and LH hormone output are increased substantially (Burger et al., 2004). Microarray based approaches of gene expression analysis have also shown only modest increases in mRNA levels in response to various condition of GnRH stimulation (Kakar et al., 2003; Lawson et al., 2007; Wurmbach et al., 2001). Increased LH protein synthesis can occur in the absence of transcriptional activity (Nguyen et al., 2004) and increased DUSP1 protein levels precedes increases in mRNA (Lawson et al., 2007; Nguyen et al., 2010), providing evidence in gonadotropes of an independent and complementary translational regulatory regime.
Earlier work showed that GnRH activates the cap-dependent translational initiation machinery and this may provide a regulatory mechanism to increase overall levels of protein synthesis to meet secretory demand (Nguyen et al., 2004). Other work has shown that GnRH induces the UPR and transiently attenuates the translation of Lhb and Cga mRNAs (Do et al., 2009). These regulatory schemes are not entirely incompatible and together may allow for fine regulation of translation that fundamentally contributes to protein quality and quantity. For example, it is known that the UPR maintains integrity of the ER and the loss of UPR signaling components compromises differentiation and the ability of the cell to manage stress (Feng et al., 2009; Saito et al., 2011; Wei et al., 2008; Zhang et al., 2002). Specifically, in this study we show that GnRH elicits a complex regulatory response that includes both enrichment of mRNA in polyribosomes or RNP subcellular fractions, suggesting both processes are integral to the GnRH regulatory response. Such a mixed response may allow for gonadotropes to increase the overall rate of translation via activation of cap-dependent initiation, select specific mRNAs for regulation via structures such as the 3′ UTR, and control the quality of protein synthesis by engaging the UPR.
It is of interest that Dusp1 is a target of translational control by GnRH. The transient activation of MAPK1/3 can be modulated by DUSP1 levels through an ultrashort feedback loop. (Caunt et al., 2008; Nguyen et al., 2010) and DUSP1 is modulated through phosphorylation by MAPK1/3, which contributes to regulation of activity or protein stability and gene transcription (Brondello et al., 1999; Jeffrey et al., 2006; Li et al., 2011). DUSPs are important modulators of the MAPK signaling and participate in establishing hysteresis of signaling cascades (Bhalla et al., 2002). The precise mechanisms for enrichment of Dusp1 mRNA in polyribosomes after GnRH stimulation remain to be fully described. Although we have identified the interaction of Dusp1 mRNA with ELAV1, an AU-rich element-binding protein, its role is not clear. Other factors interacting at the 3′UTR or elsewhere may contribute to the overall regulation of Dusp1 and similarly regulated mRNAs. Identification of other RNA binding proteins such a tristetraprolin (Clement et al., 2011) or NF90 (Kuwano et al., 2008) and definition of their role in the translational control of Dusp1 and similar mRNAs identified here may shed light on the overall mechanisms by which cells achieve targeted translational regulation of specific mRNAs. Confirmation of interaction of similarly regulated mRNAs with specific mRNA binding proteins or identification of specific RNA structures or motifs in regulated mRNA’s that confer polyribosomal enrichment will be required to fully understand the mechanisms controlling this process.
In summary, we have shown that GnRH is not equivalent to chemical insult with regard to UPR activation and its effect on Dusp1 mRNA distribution. GnRH induces a change in the mRNA composition of polyribosomes and ribonucleoprotein complexes in LβT2 cells. Evaluation of the behavior of individual mRNAs shows that the redistribution of mRNA can be attributed to both UPR and MAP kinase signaling cascades. Transcriptome-wide assessment of the compositional changes in mRNA indicates that select mRNAs are sensitive to this regulatory mechanism and are specifically enriched in either the polyribosome or RNP pools. Finally, analysis of the Dusp1 mRNA suggests that resistance to UPR-induced translational pausing may be attributed to the 3′ UTR and may involve regulation by RNA-binding proteins. Although the 3′UTR binding protein ELAV1 is associated with the Dusp1 mRNA in LβT2 cells, resistance to translational pausing requires an intact 3′UTR.
Supplementary Material
Highlights.
GnRH treatment of gonadotrope-derived cells causes remodeling of ribosome content.
Enrichment of mRNA in non-translating RNP complexes is associated with activation of EIF2AK3/PERK.
Enrichment of mRNA in active polyribosome complexes is associated with activation of ERK.
Dusp1 mRNA, a factor important for the regulatory feedback of ERK, is enriched in polyribosomes.
The mRNA binding protein ELAV1 binds the DUSP1 3′UTR and confers resistance to translational suppression induced by GNRH.
Acknowledgments
Supported by NIH Grant R01 HD 037568 and U54 HD012303 to M.A.L. M.T.D. was supported in part by NIH grant T32 GM08666. T.K. was supported in part by T32 HD007203. U.A. was supported by T36 GM095349.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarid ET, Windle JJ, Whyte DB, Mellon PL. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development. 1996;122:3319–3329. doi: 10.1242/dev.122.10.3319. [DOI] [PubMed] [Google Scholar]
- Armstrong SP, Caunt CJ, Fowkes RC, Tsaneva-Atanasova K, McArdle CA. Pulsatile and sustained gonadotropin-releasing hormone (GnRH) receptor signaling: does the Ca2+/NFAT signaling pathway decode GnRH pulse frequency? J Biol Chem. 2009;284:35746–35757. doi: 10.1074/jbc.M109.063917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SP, Caunt CJ, Fowkes RC, Tsaneva-Atanasova K, McArdle CA. Pulsatile and sustained gonadotropin-releasing hormone (GnRH) receptor signaling: does the ERK signaling pathway decode GnRH pulse frequency? J Biol Chem. 2010;285:24360–24371. doi: 10.1074/jbc.M110.115964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- Bhalla US, Ram PT, Iyengar R. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- Brondello JM, Pouyssegur J, McKenzie FR. Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science. 1999;286:2514–2517. doi: 10.1126/science.286.5449.2514. [DOI] [PubMed] [Google Scholar]
- Burger LL, Dalkin AC, Aylor KW, Haisenleder DJ, Marshall JC. GnRH pulse frequency modulation of gonadotropin subunit gene transcription in normal gonadotropes-assessment by primary transcript assay provides evidence for roles of GnRH and follistatin. Endocrinology. 2002;143:3243–3249. doi: 10.1210/en.2002-220216. [DOI] [PubMed] [Google Scholar]
- Burger LL, Haisenleder DJ, Dalkin AC, Marshall JC. Regulation of gonadotropin subunit gene transcription. J Mol Endocrinol. 2004;33:559–584. doi: 10.1677/jme.1.01600. [DOI] [PubMed] [Google Scholar]
- Burger LL, Haisenleder DJ, Marshall JC. GnRH pulse frequency differentially regulates steroidogenic factor 1 (SF1), dosage-sensitive sex reversal-AHC critical region on the X chromosome gene 1 (DAX1), and serum response factor (SRF): potential mechanism for GnRH pulse frequency regulation of LH beta transcription in the rat. Endocrine. 2011;39:212–219. doi: 10.1007/s12020-011-9440-y. [DOI] [PubMed] [Google Scholar]
- Caunt CJ, Armstrong SP, Rivers CA, Norman MR, McArdle CA. Spatiotemporal regulation of ERK2 by dual specificity phosphatases. J Biol Chem. 2008;283:26612–26623. doi: 10.1074/jbc.M801500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Do AN, Kimball SR, Cavener DR, Jefferson LS. eIF2alpha kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol Genomics. 2009;38:328–341. doi: 10.1152/physiolgenomics.90396.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A, Jaffrey SR. Insights into the roles of local translation from the axonal transcriptome. Open Biol. 2012;2:120079. doi: 10.1098/rsob.120079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do MH, Santos SJ, Lawson MA. GnRH induces the unfolded protein response in the LbetaT2 pituitary gonadotrope cell line. Molecular endocrinology. 2009;23:100–112. doi: 10.1210/me.2008-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobkin-Bekman M, Rahamin-Ben Navi L, Shterntal B, Sviridonov L, Przedecki F, Naidich-Exler M, Brodie C, Seger R, Naor Z. Differential role of PKC isoforms in GnRH and phorbol 12-myristate 13-acetate activation of extracellular signal-regulated kinase and Jun N-terminal kinase. Endocrinology. 2010;151:4894–4907. doi: 10.1210/en.2010-0114. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons J, Townley-Tilson WH, Deleault KM, Skinner SJ, Gross RH, Whitfield ML, Brooks SA. Identification of TTP mRNA targets in human dendritic cells reveals TTP as a critical regulator of dendritic cell maturation. RNA. 2008;14:888–902. doi: 10.1261/rna.748408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Wei J, Gupta S, McGrath BC, Cavener DR. Acute ablation of PERK results in ER dysfunctions followed by reduced insulin secretion and cell proliferation. BMC Cell Biol. 2009;10:61. doi: 10.1186/1471-2121-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris HA, Shupnik MA. Mechanisms for pulsatile regulation of the gonadotropin subunit genes by GNRH1. Biol Reprod. 2006;74:993–998. doi: 10.1095/biolreprod.105.049049. [DOI] [PubMed] [Google Scholar]
- Fribley A, Zhang K, Kaufman RJ. Regulation of apoptosis by the unfolded protein response. Methods Mol Biol. 2009;559:191–204. doi: 10.1007/978-1-60327-017-5_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Devine T, George AD, Chittur SV, Baroni TE, Penalva LO, Tenenbaum SA. RIP-Chip analysis: RNA-Binding Protein Immunoprecipitation-Microarray (Chip) Profiling. Methods Mol Biol. 2011;703:247–263. doi: 10.1007/978-1-59745-248-9_17. [DOI] [PubMed] [Google Scholar]
- Jeffrey KL, Brummer T, Rolph MS, Liu SM, Callejas NA, Grumont RJ, Gillieron C, Mackay F, Grey S, Camps M, Rommel C, Gerondakis SD, Mackay CR. Positive regulation of immune cell function and inflammatory responses by phosphatase PAC-1. Nat Immunol. 2006;7:274–283. doi: 10.1038/ni1310. [DOI] [PubMed] [Google Scholar]
- Kakar SS, Winters SJ, Zacharias W, Miller DM, Flynn S. Identification of distinct gene expression profiles associated with treatment of LbetaT2 cells with gonadotropin-releasing hormone agonist using microarray analysis. Gene. 2003;308:67–77. doi: 10.1016/s0378-1119(03)00446-3. [DOI] [PubMed] [Google Scholar]
- Kondratyev M, Avezov E, Shenkman M, Groisman B, Lederkremer GZ. PERK-dependent compartmentalization of ERAD and unfolded protein response machineries during ER stress. Exp Cell Res. 2007;313:3395–3407. doi: 10.1016/j.yexcr.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Kuhn KM, DeRisi JL, Brown PO, Sarnow P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol Cell Biol. 2001;21:916–927. doi: 10.1128/MCB.21.3.916-927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson MA, Tsutsumi R, Zhang H, Talukdar I, Butler BK, Santos SJ, Mellon PL, Webster NJ. Pulse sensitivity of the luteinizing hormone beta promoter is determined by a negative feedback loop Involving early growth response-1 and Ngfi-A binding protein 1 and 2. Molecular endocrinology. 2007;21:1175–1191. doi: 10.1210/me.2006-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc GM, Boockfor FR, Faught WJ, Frawley LS. Development of a destabilized firefly luciferase enzyme for measurement of gene expression. Biotechniques. 2000;29:590–591. 594–596, 598. doi: 10.2144/00293rr02. passim. [DOI] [PubMed] [Google Scholar]
- Li S, Zhu F, Zykova T, Kim MO, Cho YY, Bode AM, Peng C, Ma W, Carper A, Langfald A, Dong Z. T-LAK cell-originated protein kinase (TOPK) phosphorylation of MKP1 protein prevents solar ultraviolet light-induced inflammation through inhibition of the p38 protein signaling pathway. J Biol Chem. 2011;286:29601–29609. doi: 10.1074/jbc.M111.225813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Pnueli L, Tan JH, Naor Z, Rajagopal G, Melamed P. Negative feedback governs gonadotrope frequency-decoding of gonadotropin releasing hormone pulse-frequency. PLoS One. 2009;4:e7244. doi: 10.1371/journal.pone.0007244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin NY, Lin CT, Chang CJ. Modulation of immediate early gene expression by tristetraprolin in the differentiation of 3T3-L1 cells. Biochem Biophys Res Commun. 2008;365:69–74. doi: 10.1016/j.bbrc.2007.10.119. [DOI] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JWB. Origins and Principles of Translational Control. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. pp. 1–31. [Google Scholar]
- Nanbru C, Lafon I, Audigier S, Gensac MC, Vagner S, Huez G, Prats AC. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- Nguyen KA, Intriago RE, Upadhyay HC, Santos SJ, Webster NJ, Lawson MA. Modulation of gonadotropin-releasing hormone-induced extracellular signal-regulated kinase activation by dual-specificity protein phosphatase 1 in LbetaT2 gonadotropes. Endocrinology. 2010;151:4882–4893. doi: 10.1210/en.2009-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KA, Santos SJ, Kreidel MK, Diaz AL, Rey R, Lawson MA. Acute regulation of translation initiation by gonadotropin-releasing hormone in the gonadotrope cell line LbetaT2. Molecular endocrinology. 2004;18:1301–1312. doi: 10.1210/me.2003-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld S, Wang T, Kim Y, Milner J. Numerical deconvolution of cDNA microarray signal: simulation study. Ann NY Acad Sci. 2004;1020:110–123. doi: 10.1196/annals.1310.012. [DOI] [PubMed] [Google Scholar]
- Ruf F, Sealfon SC. Genomics view of gonadotrope signaling circuits. Trends Endocrinol Metab. 2004;15:331–338. doi: 10.1016/j.tem.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Saito A, Ochiai K, Kondo S, Tsumagari K, Murakami T, Cavener DR, Imaizumi K. Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem. 2011;286:4809–4818. doi: 10.1074/jbc.M110.152900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev. 2008;29:317–333. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J Roy Stat Soc Ser B. 2002:479–498. [Google Scholar]
- Tsaneva-Atanasova K, Caunt CJ, Armstrong SP, Perrett RM, McArdle CA. Decoding neurohormone pulse frequency by convergent signalling modules. Biochem Soc Trans. 2012;40:273–278. doi: 10.1042/BST20110645. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilyev VV, Lawson MA, Dipaolo D, Webster NJ, Mellon PL. Different signaling pathways control acute induction versus long-term repression of LHbeta transcription by GnRH. Endocrinology. 2002;143:3414–3426. doi: 10.1210/en.2001-211215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verfaillie T, Rubio N, Garg AD, Bultynck G, Rizzuto R, Decuypere JP, Piette J, Linehan C, Gupta S, Samali A, Agostinis P. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 2012 doi: 10.1038/cdd.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wei J, Sheng X, Feng D, McGrath B, Cavener DR. PERK is essential for neonatal skeletal development to regulate osteoblast proliferation and differentiation. J Cell Physiol. 2008;217:693–707. doi: 10.1002/jcp.21543. [DOI] [PubMed] [Google Scholar]
- Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. Gonadotropin-releasing hormone receptor-coupled gene network organization. J Biol Chem. 2001;276:47195–47201. doi: 10.1074/jbc.M108716200. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. Protein folding in the endoplasmic reticulum and the unfolded protein response. Handb Exp Pharmacol. 2006:69–91. doi: 10.1007/3-540-29717-0_3. [DOI] [PubMed] [Google Scholar]
- Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Roberson MS. Role of MAP kinase phosphatases in GnRH-dependent activation of MAP kinases. J Mol Endocrinol. 2006;36:41–50. doi: 10.1677/jme.1.01881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.