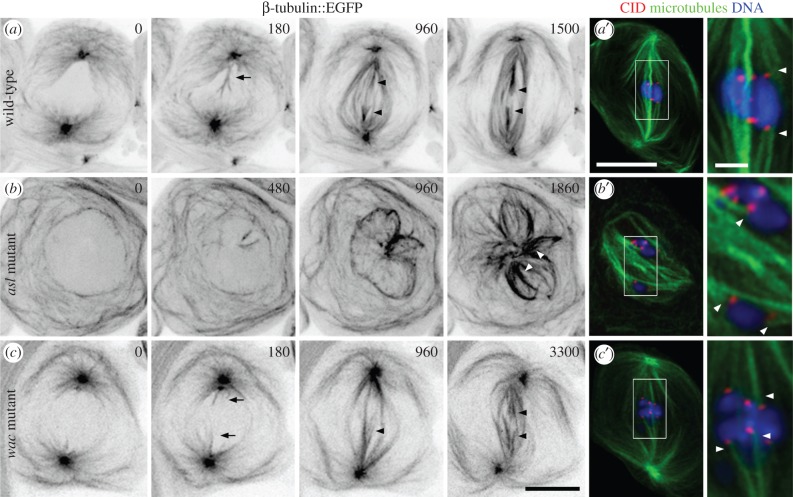
Figure 1.
Meiotic spindles form through centrosomal, acentrosomal and Wac-mediated pathways. (a–c) Time-lapse sequences of spindle formation in the indicated genetic backgrounds visualized using β-tubulin::EGFP expression. (a) Wild-type spindle formation initiates as centrosome-nucleated astral MTs penetrate into the nucleus (180; arrow). These increase in number and organize into bundles (180–960), some of which are k-fibres (arrowheads) as revealed by their contact with chromosome-based fluorescence ‘ghosts’. The chromosomes align at the spindle equator where they remain at metaphase (1500), shortly before anaphase entry. (a′) Fixed wild-type metaphase cell showing the distribution of the centromeric protein CID/CENP-A as a marker for kinetochore position, MTs and DNA. Arrowheads denote k-fibres. (b) A centrosome inactivated, asl2/asl3 mutant. Neither centrosomes nor asters are detected. Spindle assembly is first observed with the nucleation of a few MTs within the nucleus. Progressively more MTs appear that interact (480–960) to establish larger structures. These are poorly organized, but exhibit k-fibre-like MT bundles (1860; arrowheads). (b′) Fixation and staining of asl2/asl3 cells confirms that k-fibres (arrowheads) form in the absence of centrosomes. (c) Spindle assembly in a wac Δ12 hemizygous mutant. Like the wild-type, centrosomal MTs invade the nucleus (0–180; arrows). However, these require protracted periods to organize into a spindle with recognizable albeit non-robust k-fibres (960; arrowhead). As the k-fibres mature (arrowheads) the chromosomes assume an equatorial position where they remain throughout metaphase (3300). (c′) Fixation and staining of wac mutant cells confirms that this protein and by extension the Augmin complex are not needed for k-fibre formation (arrowheads) or normal spindle morphology. Time is in seconds relative to the onset of spindle formation. All images are z-projections. Bars are 10 μm except in zoomed fixed images where they are 2 μm.