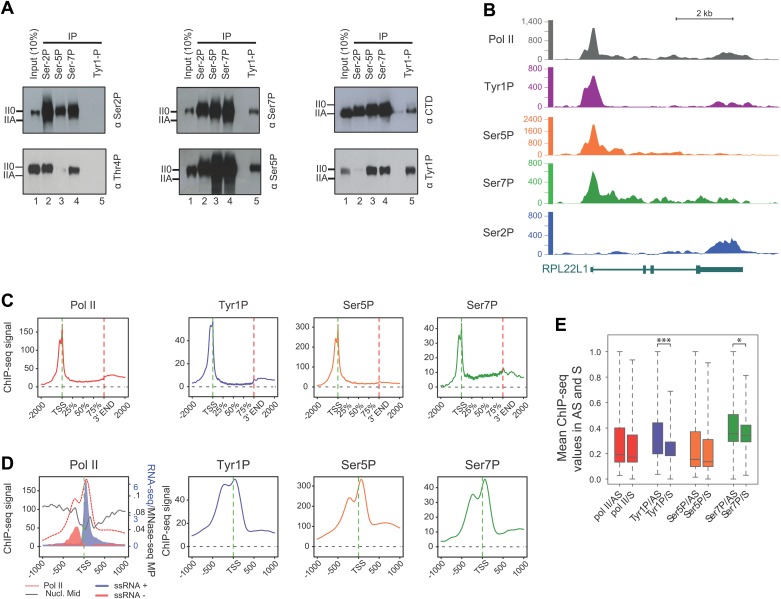
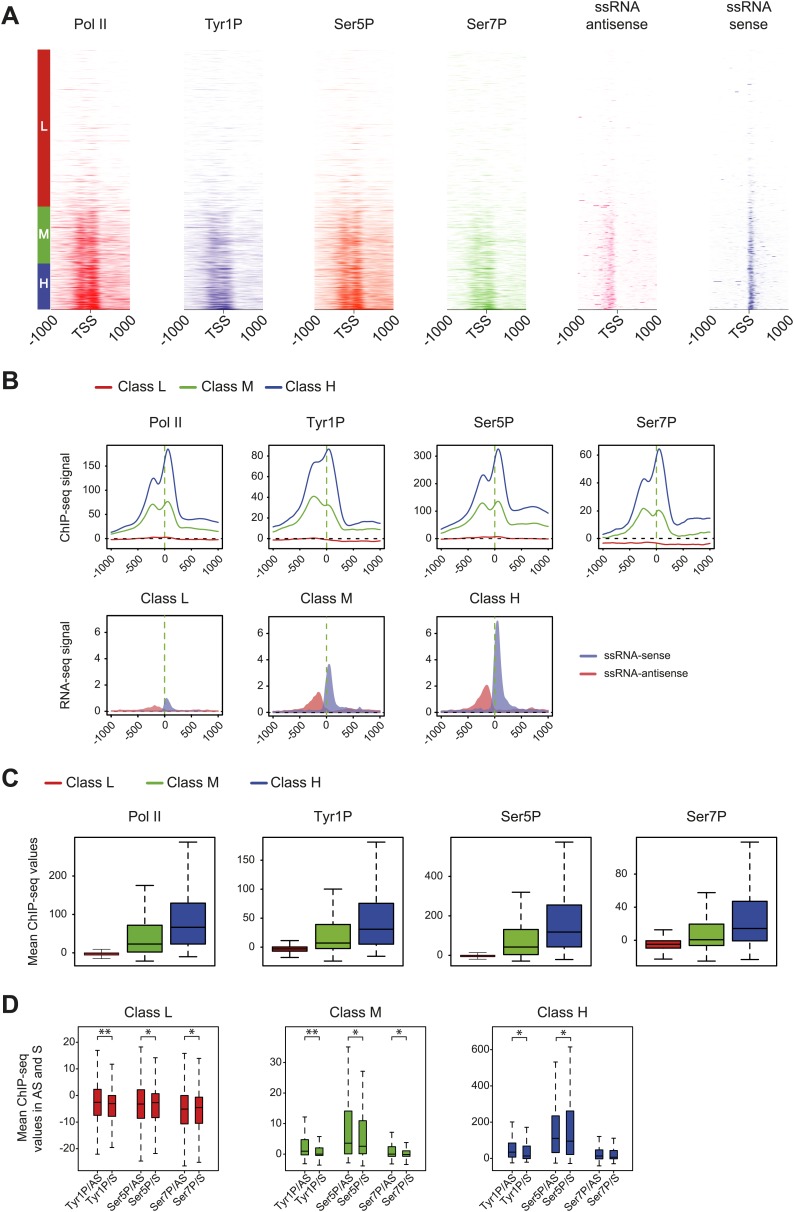
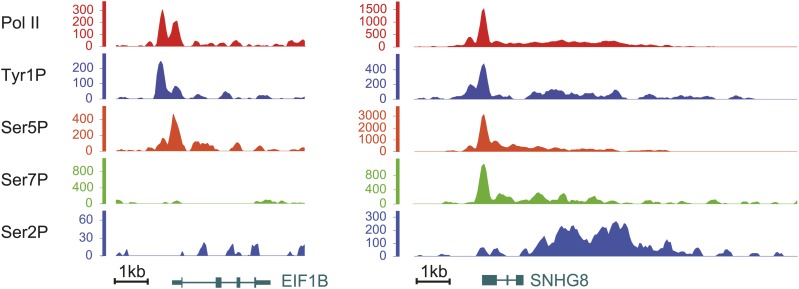
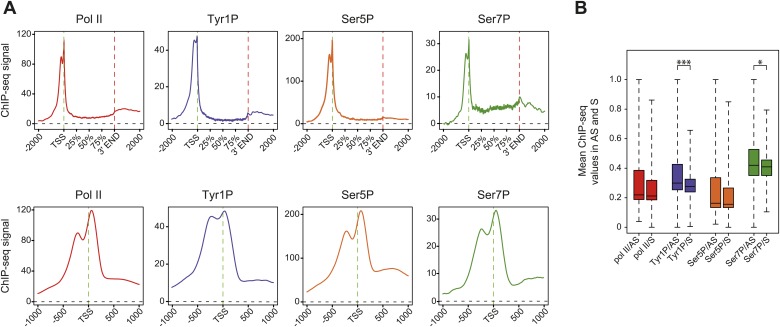
Figure 2. CTD Tyrosine 1 is phosphorylated mainly at TSS and is dominant in antisense transcription.
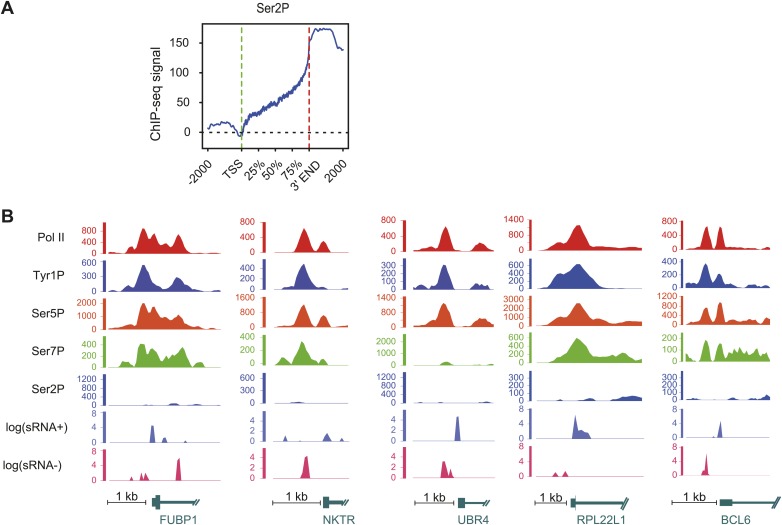
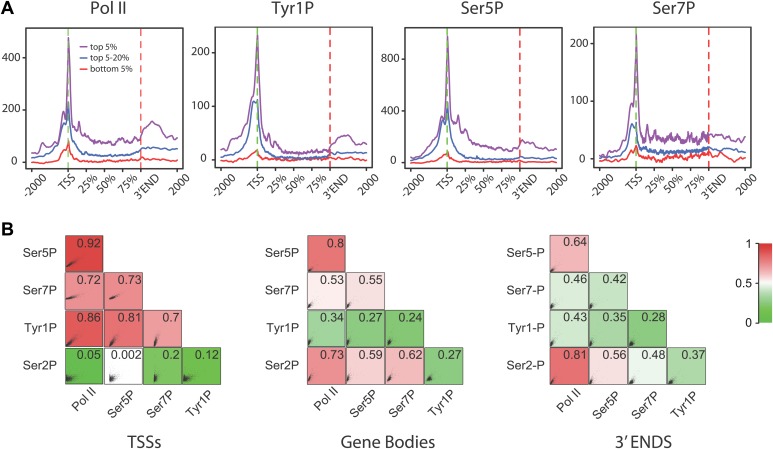
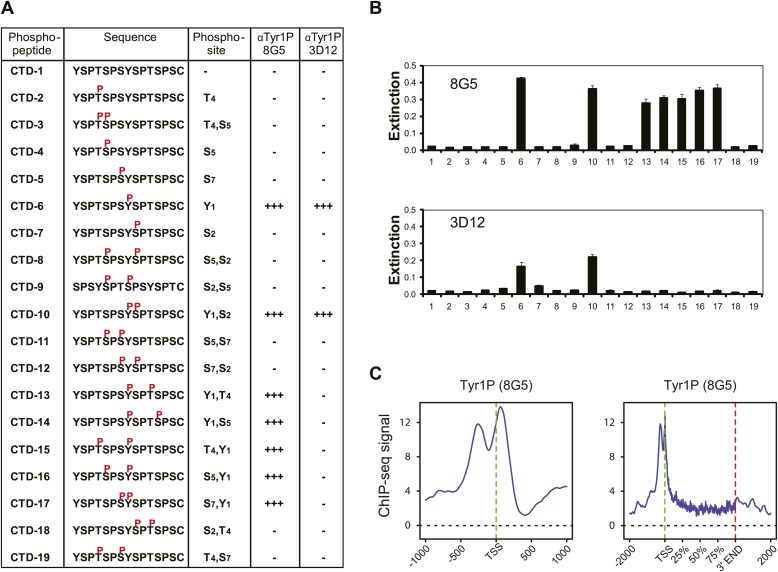
(A) Co-immunoprecipitation with specific CTD isoforms in Raji B-cells reveals Tyr1P (3D12) association with Ser5P and Ser7P but not with Ser2P and Thr4P. (B) ChIP-seq example illustrating Tyr1P (3D12) association around the promoter of RPL22L1 gene. (C) Composite average profiling of ChIP-seq data at coding genes locations for Pol II (1433 genes), Tyr1P (3D12, 2462 genes), Ser5P (1464 genes), and Ser7P (2186 genes) in Raji B-cells and based on selections described in Figure 2—figure supplement 1B. Less stringent selections with more genes gave equivalent profiling (Figure 2—figure supplement 4A). (D) Profiling of Pol II, Tyr1P (3D12), Ser5P, Ser7P, nucleosomes midpoint and short strand specific RNAs (ssRNAs) around TSS locations with same selections described in (C). (E) Boxplots on 3201 genes without outliers showing mean levels of Pol II (2986 genes), Tyr1P (2964 genes), Ser5P (2909 genes), and Ser7P (2948 genes) ChIP-seq signal on regions representing each transcription orientation. The p-values (parametric two sided paired t test) of the difference of AS vs S signal are Pol II = 0.5, Tyr1p=3.4 × 10−15, Ser5p=0.6, Ser7p=3.5 × 10−2.