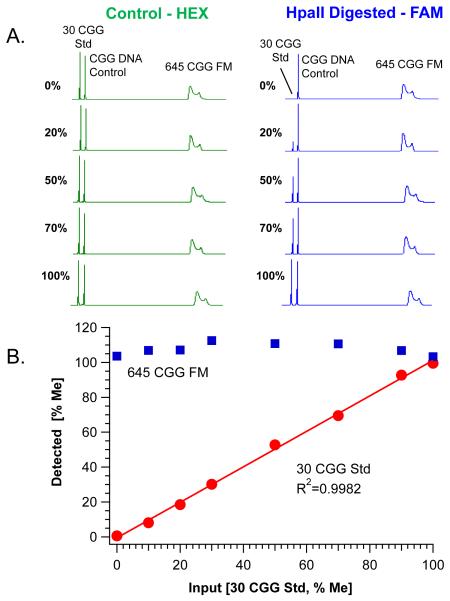
Fig. 2. Methylation PCR accurately and reproducibly recovers the proportion of methylated DNA standards.
A) Electropherograms of PCR products derived from the Control (HEX, left) and HpaII digested (FAM, right) reactions of methylation standards ranging from 0 to 100% methylation are shown. Each reaction included a variable input of methylation standard with a constant concentration of CGG DNA Control and 645 CGG full mutation (FM) allele from cell line DNA (NA04025). Product peaks amplified from the control digestion are shown relative to corresponding percent methylation of the 30 CGG plasmid (30 CGG Std). B) Plot of the linear fit of known input methylated DNA standards versus detected percent methylation. Quantification of the background, fully methylated 645 CGG allele is superposed (Mean=108±3%) across each methylation standard.