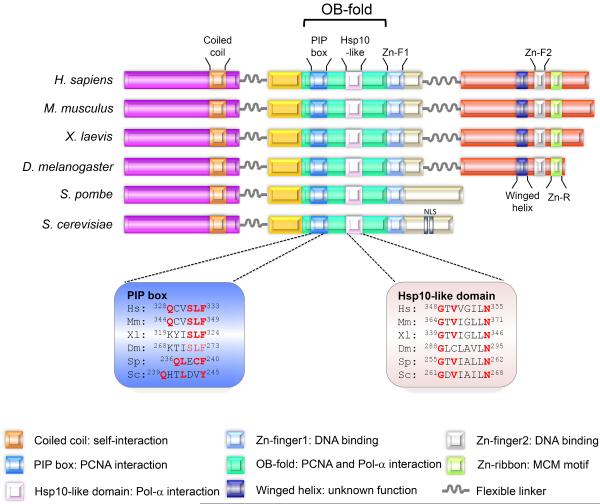
Figure 1. Structural architecture of Mcm10.
The functional domains of Mcm10 across different species are shown. The coiled-coil motif within the NTD mediates Mcm10 oligomerization. The ID includes a PIP box and Hsp10-like domain, both of which reside in the OB-fold. The PIP box and the Hsp10-like domain mediate Mcm10's interaction with PCNA and Pol-α, respectively, and sequence alignments of these regions are shown. (The positions of the indicated amino acids within these domains for HsMcm10 are in reference to isoform 1, consistent with the cancer mutations described in Figure 5a). The OB-fold and Zn-F1 provide a binding platform for DNA. Metazoan Mcm10 harbors additional DNA- and Pol-α-binding regions within the CTD. DNA binding within the CTD is primarily mediated by Zn-F2. How the Zn-R and winged helix motif contribute to protein function is currently unknown. Nuclear localization sequences (NLS) have only been identified in S. cerevisiae. Mcm10 orthologs are illustrated for Hs: Homo sapiens; Mm: Mus musculus, Xl: Xenopus laevis, Dm: Drosophila melanogaster, Sp: Schizosaccharomyces pombe, and Sc: Saccharomyces cerevisiae.