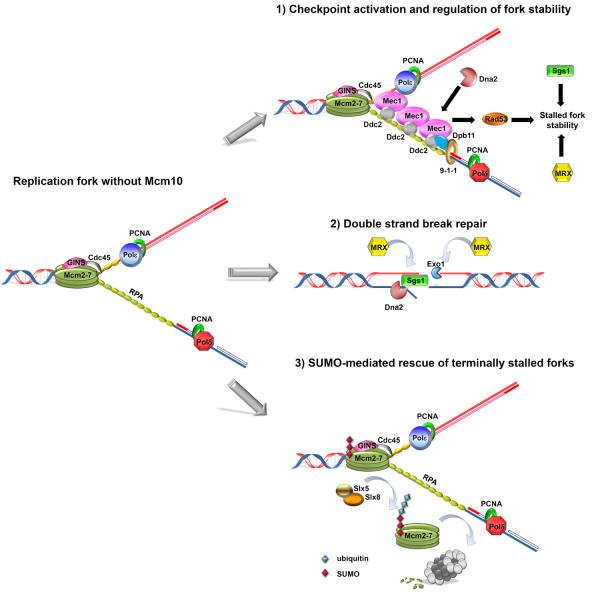
Figure 4. Proposed molecular mechanisms that alleviate replication stress following Mcm10 depletion.
On the left, a replication fork is illustrated in the absence of Mcm10. We hypothesize that Pol-α is recruited less efficiently to chromatin and that this defect is more pronounced on the lagging strand template. These events lead to the accumulation of RPA-coated ssDNA. Based on the genetic interaction, we propose that Mcm10 depletion triggers three possible scenarios: 1) Stretches of RPA-coated ssDNA are recognized by Mec1:Ddc2 complexes, initiating checkpoint activation. This response is enhanced by the 9-1-1 checkpoint clamp (Rad9-Hus1-Rad1), which binds to the 5' recessed ds RNA/DNA hybrid formed by an Okazaki fragment and the lagging strand template. The 9-1-1 clamp recruits Dpb11, which subsequently activates Mec1. In addition to Dpb11, Dna2 can also activate Mec1. Mec1 phosphorylates Rad53, leading to fork stabilization. Sgs1 and the MRX complex can also execute this function independently of the checkpoint. 2) Failure to stabilize the stalled fork in mcm10 mutants results in fork collapse and DSBs. DSB repair is initiated when the MRX complex and Sae2 mediate short-range resection of dsDNA ends. After this step, the MRX complex recruits Dna2, Sgs1/Top3/Rmi1 and Exo1, which perform additional resection to generate products compatible with homologous recombination (Sae2, Top3 and Rmi1 are not shown). 3) Possible mechanism to promote DSB formation in mcm10 mutants through Slx5/Slx8-mediated fork collapse. The Slx5/Slx8 complex ubiquitinates SUMOylated proteins. A hypothetical target shown here is the Mcm2-7 complex, which is degraded by the proteasome (shown in light and dark grey). Proteolysis results in disassembly of the replisome and collapse of terminally stalled forks.