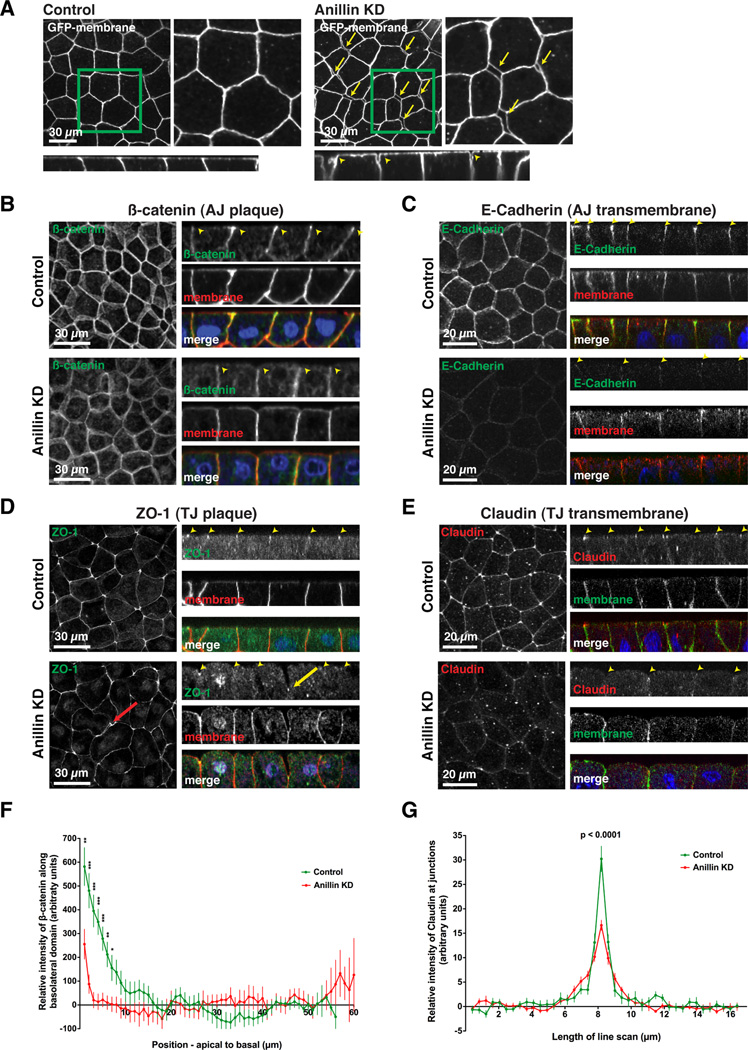
Figure 2. Adherens junctions and tight junctions are disrupted when Anillin is knocked down.
A. Single intermediate plane views (top) and z views (bottom) of GFP-membrane in control and Anillin KD embryos reveal increased intercellular spaces in Anillin KD embryos (yellow arrows and arrowheads).
B-E. Fixed staining of control and Anillin KD embryos for β-catenin (B), E-Cadherin (C), ZO-1 (D), and Claudin (E). GFP-mem or mChe-mem was used as a MO injection marker, and DAPI labels DNA. Z views show the normal localization of the cell-cell junction proteins in control cells as well as the disrupted localization in Anillin KD cells (see yellow arrowheads). The x-y tight junction protein images on the left in D and E are maximal intensity projections of serial z sections. The red arrow in D highlights an intercellular space between a dividing cell and its neighbor, while the yellow arrow indicates a ZO-1 concentration that is buried basally.
F. Quantification of β-catenin polarization in control and Anillin KD cells from line scans along the basolateral surface. The β-catenin signal at the 10 most basal points was normalized to zero so that data from multiple embryos could be averaged (see Experimental Procedures for details). Data is from two independent experiments, n = 26 embryos for control, n = 18 embryos for Anillin KD, graphed as mean +/− SEM, * indicates p ≤ 0.05, ** indicates p ≤ 0.01, *** indicates p ≤ 0.001.
G. Quantification of the relative intensity of Claudin at cell-cell junctions by generating line scans perpendicular to junctions (see Experimental Procedures for details). Data is from two independent experiments, n = 10 embryos for control, n = 12 for Anillin KD, graphed as mean +/− SEM, p < 0.0001.
See also Figure S2.