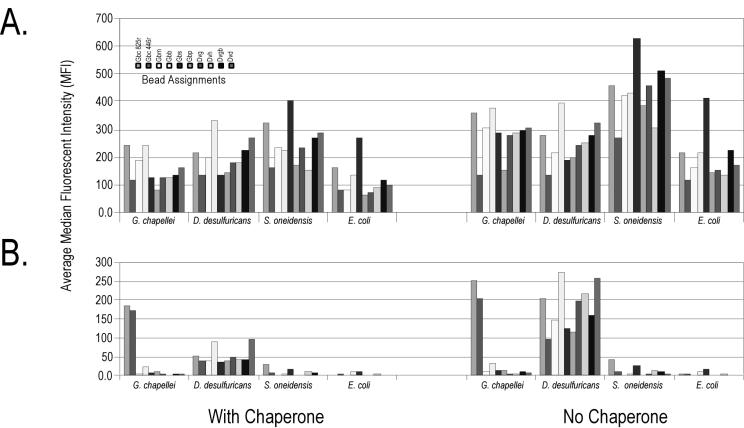
FIG. 2.
Achieving hybridization specificity on a tunable surface suspension array during manual nucleic acid hybridization. A sample of 100 ng of target RNA was heat denatured in the presence of 1 μg of salmon sperm DNA with or without a 250 nM concentration of each of the two chaperone probes in pH 5 HS phosphate buffer. Hybridization reactions proceeded for 2 h at room temperature. Beads then were washed with 2× SSC-0.02% Tween 20-0.5% Sarkosyl (pH 7) buffer at room temperature before buffer exchange into 1× SSC-0.01% Tween 20-0.25% Sarkosyl (pH 7) and analysis on the flow cytometer. (A) Intact RNA targets. (B) Fragmented RNA targets. Data are corrected for background, and error bars are omitted for presentation clarity. The order of the bead assignments (from left to right) in the key is the same as in the bar graphs.