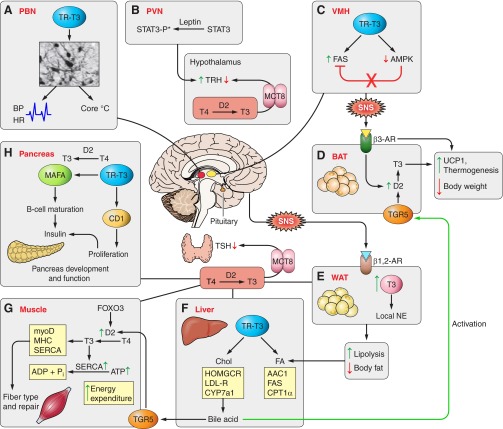
FIGURE 1.
Overview of sites of thyroid hormone regulation of metabolism. Hypothalamic-Pituitary-Thyroid axis: thyrotropin releasing hormone (TRH) and thyroid stimulating hormone (TSH) respond primarily to circulating serum T4, converted in the hypothalamus and pituitary to T3 by the 5′-deiodinase type 2 (D2). The monocarboxylate transporter 8 (MCT8) is required for T3 transport into the pituitary and hypothalamus. A, parvalbuminergic neurons (PBN): PBN are a population of newly discovered neurons in the anterior hypothalamus that are directly linked to the regulation of cardiovascular function, including heart rate, blood pressure, and body temperature. Thyroid hormone receptor signaling is required for the normal development of PBN neurons linking thyroid hormone to cardiac and temperature regulation. B, paraventricular nucleus of the hypothlamus (VPN): leptin, produced in peripheral fat tissue, provides feedback at the VPN, stimulates signal transducer and activator of transcription (STAT)3 phosphorylation (STAT3-P*), which directly stimulates TRH expression. Leptin also stimulates TRH indirectly in the arcuate nucleus by inhibiting neuropeptide Y and agouti-related protein, stimulating proopiomelanocortin (POMC), and the POMC product α-melanocyte stimulating hormone (α-MSH) stimulates CREB in the TRH neuron (indirect pathway is not shown in Figure 1). C, ventromedial nucleus of the hypothalamus (VMH): hyperthyroidism or T3 treatment stimulates de novo fatty acid synthesis in the VMH, which inhibits AMPK phosphorylation and increases fatty acid synthase (FAS) activity. Increased hypothalamic lipid synthesis is associated with activation of the sympathetic nervous system (SNS) which stimulates brown adipose tissue (BAT). D, BAT: adrenergic signaling through the β3-adrenergic receptor (AR) stimulates UCP1 gene expression, stimulates D2 activity by deubiquitination, and promotes thermogenesis and weight loss. The metabolic signal from bile acid via the G protein-coupled membrane bile acid receptor (TGR5) has been shown in one model to stimulate D2 activity and local T3 production, which further stimulates BAT lipolysis, UCP1 expression, and thermogenesis. E, white adipose tissue (WAT): SNS signals via β1- and β2-AR stimulate WAT lipolysis. T3 stimulates local production of norepinephrine (NE), increasing lipolysis and reducing body fat. F, liver: T3 is involved in both cholesterol and fatty acid metabolism (see details in Figure 3). HOMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase; ACC1, acetyl-CoA carboxylase 1; CYP7a1, cytochrome P-450 7A1; CPT-1α, carnitine palmitoyltransferase 1α; LDL-R, low-density lipoprotein receptor. G, muscle: Forkhead box O3 (FoxO3) induces D2 expression, increases local T3 in skeletal muscle, and promotes T3-target gene expression; myoD, myosin heavy chain (MHC) and sarcoplasmic reticulum Ca2+-ATPase (SERCA). Local T3 also determines the relative expression level of MHC and SERCA isoforms. Expression level of these isoforms determines muscle fiber types and initiation of repair. SERCA2a is primarily expressed in slow-twitch fibers and SERCA1 in fast-twitch fibers. T3 stimulates SERCA, which hydrolyzes ATP and increases energy expenditure. H, pancreas: T3 and TR are required for normal pancreatic development and function. In rat pancreatic β cells, expression of TR and D2 are activated during normal development. T3 treatment enhances Mafa (v-maf musculoaponeurotic fibrosarcoma oncogene homolog A) transcription factor gene expression and increases MAFA protein content, the key factor for maturation of β cells to secrete insulin in response to glucose. T3 stimulates cyclin D1 (CD1) gene expression and protein level and promotes proliferation. Increasing cyclin D1 activates the cyclin D1/cyclin-dependent kinase/retinoblastoma protein/E2F pathway.