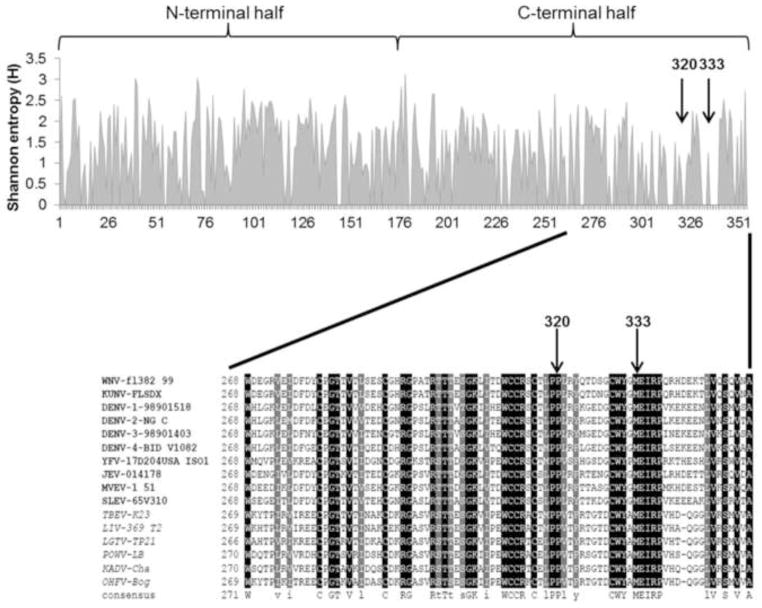
Figure 5. Shannon entropy (H) and multiple protein alignment of the NS1 proteins of several mosquito- and tick-borne flaviviruses.
The amino acid sequences of 10 mosquito-borne and 6 tick-borne flaviviruses were aligned with ClustalW using the Blosum scoring matrix. The multiple-alignment file was displayed with BOXshade 3.21 where black-shading represents identical residues and gray-shading represents conserved substitutions. Shannon entropy analysis of the multiple protein alignment was performed by the Protein Variability Server (PVS) at the Complutense University of Madrid, and the data generated was displayed in MS Excel. H ≤ 1 are considered highly conserved residues. Mosquito-borne flaviviruses are represented in bolded font and tick-borne flaviviruses in italics.