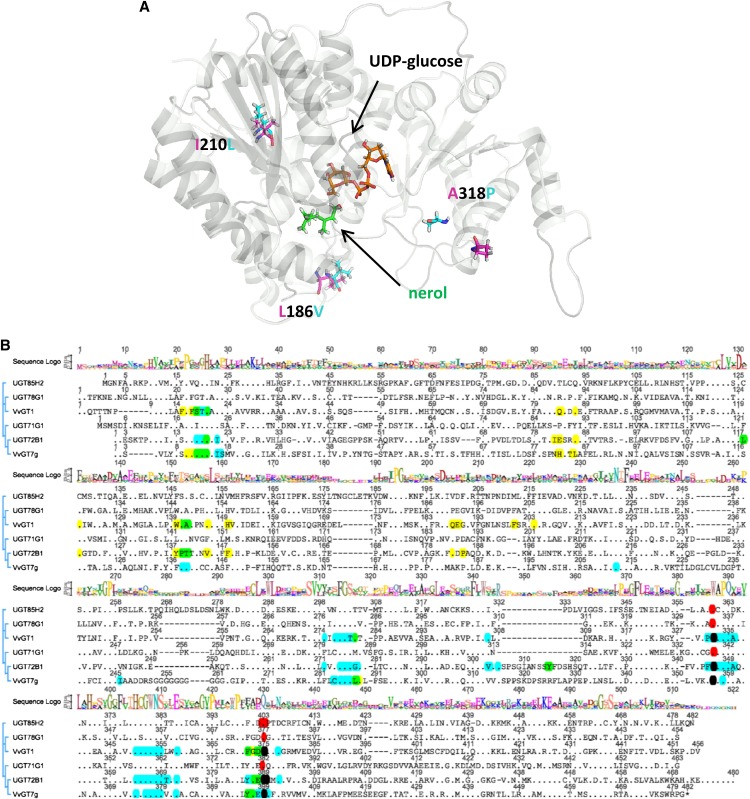
Figure 7.
Ribbon diagram of the calculated 3D structure of VvGT7b with UDP-Glc (orange) and nerol (green; A) and amino acid sequence alignment of VvGT7g and five plant GTs that have been crystallized and their 3D structures solved (B). UDP-Glc and nerol are shown as sticks and marked with arrows. β-Strands and α-helices are shown in gray. The residues at positions 186, 210, and 318, which differentiated the active from the inactive forms, are shown as sticks and are denominated. The alignment was performed using ClustalX. The plant GTs include M. truncatula UGT71G1 (protein data bank identification 2ACW), UGT78G1 (3HBF), and UGT85H2 (2PQ6), grape VvGT1 (2C1Z), and Arabidopsis UGT72B1 (2VCE). A consensus sequence logo is shown in the top traces, conserved amino acids are displayed as dots, and amino acids within 5 Å to the acceptor and sugar donor molecules (UDP-Glc) are marked in yellow and blue, respectively. Overlaps are shown in green (blue + yellow) and black (blue + red). The plant secondary glucosyltransferase box is confined by the amino acids marked in red.