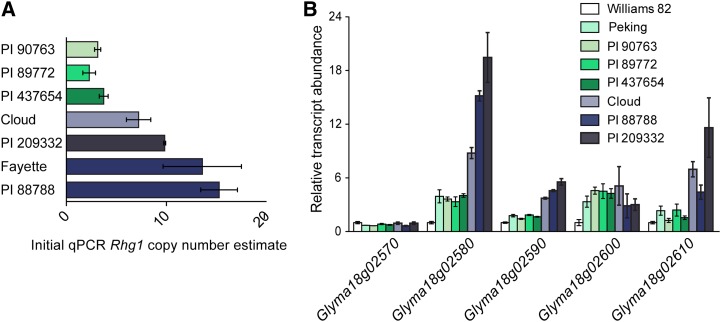
Figure 1.
Estimates of copy number and transcript abundance suggest different types of Rhg1 loci. A, Initial Rhg1 copy number estimates, obtained using qPCR to amplify genomic DNA, identify two groups of SCN-resistant lines: low-copy number, between two and four repeats (PI 90763, PI 89772, and PI 437654), and high-copy number, estimated to have greater than seven repeats (Cloud, PI 209332, Fayette, and PI 88788). Copy number is expressed as the ratio of qPCR template abundance estimates for Rhg1 repeat junction and for a nonduplicated neighboring gene. B, Transcript abundance, relative to SCN-susceptible Williams 82, indicates the presence of two expression groups of Rhg1 loci in SCN-resistant lines. Roots from lines identified in subsequent work as having three copies of the 31-kb Rhg1 repeat (Peking, PI 90763, PI 89772, and PI 437654) exhibit lower transcript abundance than lines with seven, nine, or 10 Rhg1 copies (Cloud, PI 88788, and PI 209332). The one complete copy of Glyma18g02570, located immediately outside of the Rhg1 repeat region (Cook et al., 2012; this work), is expressed at a similar level across all the tested lines. The expression level of Glyma18g02600 is near the detection limit for qPCR.