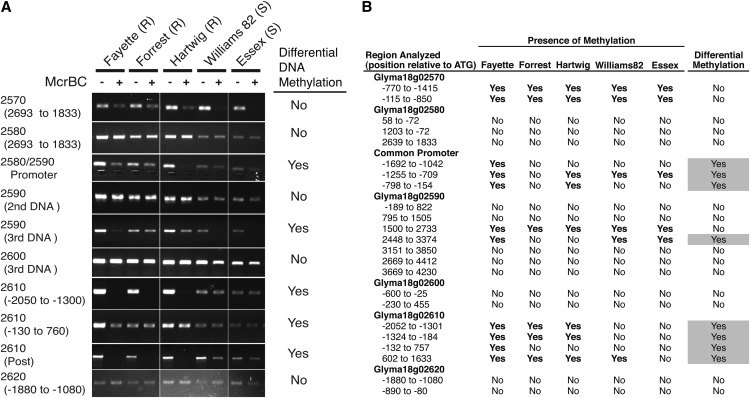
Figure 7.
Differential Rhg1 locus DNA methylation between SCN-resistant and SCN-susceptible lines, particularly in control regions upstream of SCN resistance genes. A, Representative gel images of PCR products from a soybean root genomic DNA template treated with the restriction endonuclease McrBC (+) or buffer only (−) prior to PCR. McrBC cleaves (G/A)mC sites containing methylcytosine, preventing PCR amplification of cleaved template strands so that PCR product abundance goes down with increasing levels of methylation. Differential DNA Methylation was scored as positive if any soybean line differed from other lines in McrBC sensitivity of the PCR product in two independent tests. Soybean lines are denoted as either resistant (R) or susceptible (S) to SCN. B, Summary table for the replicated McrBC study described in A with 23 PCR primer pairs used to assess DNA methylation within the Rhg1 locus. The presence of methylation is listed as yes if both DNA samples showed reduced PCR amplification following McrBC DNA treatment. The right column reports methylation differences between different soybean lines.