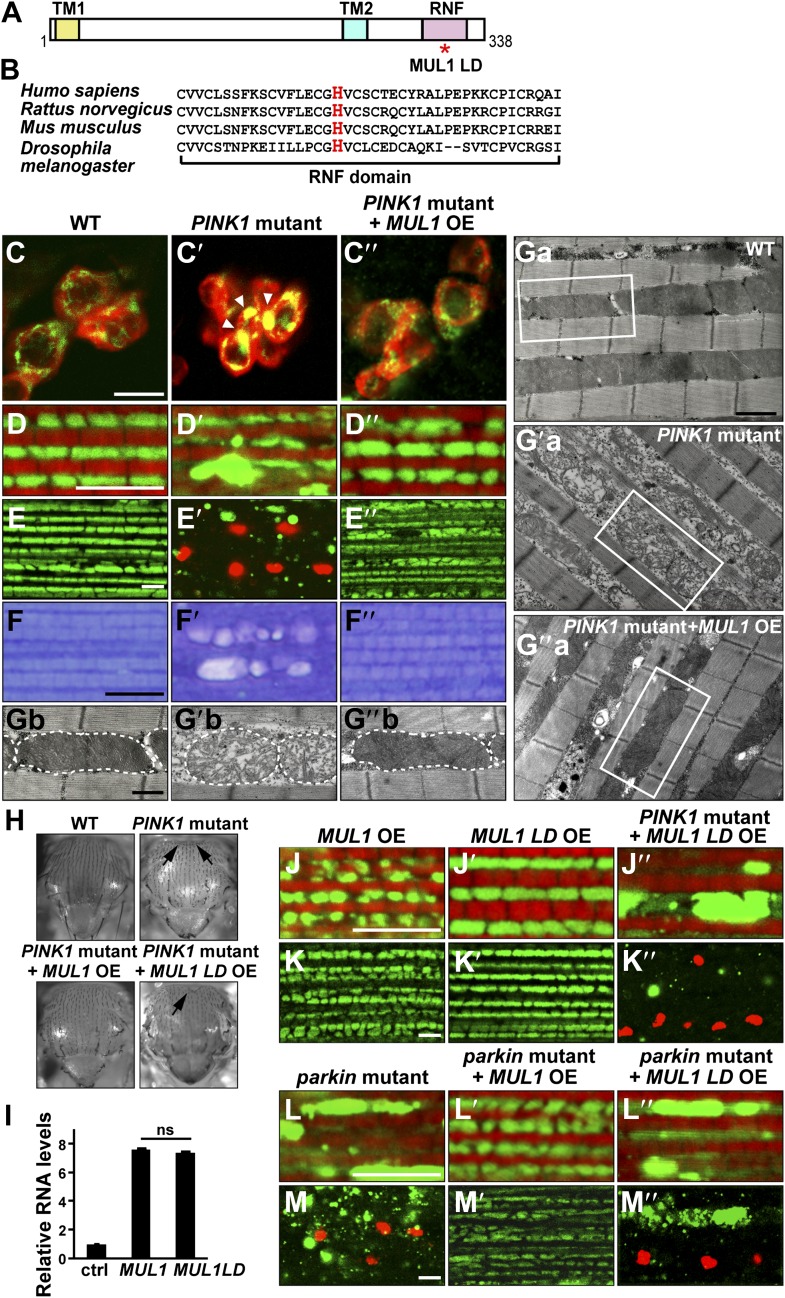
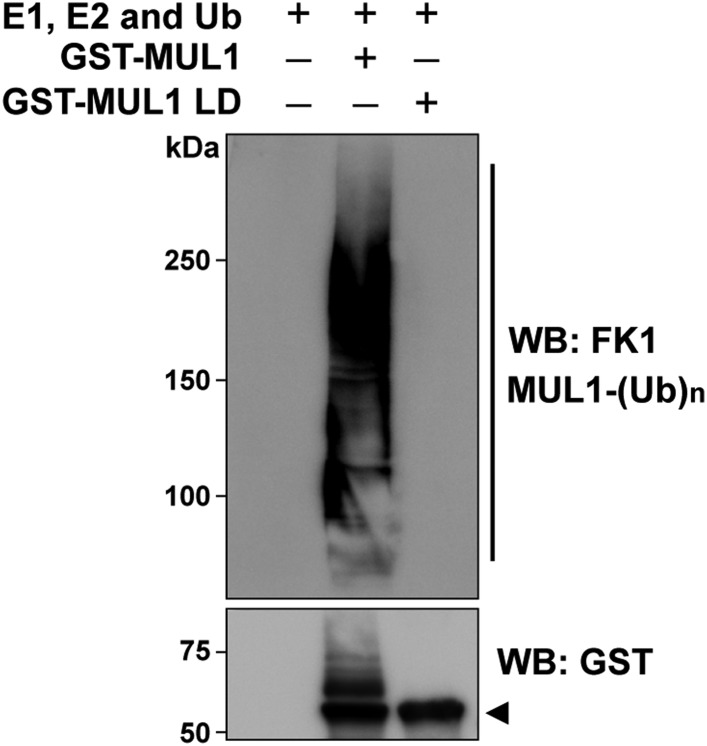
Figure 1. Overexpression of MUL1, but not MUL1 LD, suppresses PINK1/parkin mutant phenotypes.
(A) Protein domain organization of Drosophila MUL1. TM1, TM2, and RNF represent transmembrane domains 1 and 2, and the RING Finger domain, respectively. The position of the mutation in the ligase dead (LD) version of MUL1 is marked with a red asterisk. (B) Sequence alignment of MUL1 in various species in the highly conserved RNF domain. A highly conserved histidine residue (marked as red) was mutated to alanine in MUL1 LD, ablating ligase activity. (C–C″) Dopaminergic neurons stained with an anti-TH antibody in red and mitochondria labeled with mitoGFP in green. Neurons in the PPL1 cluster are shown. While mitochondria in wild-type dopaminergic neurons are dispersed (C), mitochondria in PINK1 mutant dopaminergic neurons are clumped (C′, white arrow heads). This phenotype is suppressed by MUL1 overexpression driven by TH-Gal4 (C″). Scale bars: 10 µm. (D–E″’’ and J–M″’’) Confocal images of the IFM from thoraces double labeled with mitoGFP and phalloidin (red) (D–D″, J–J″, L–L″), or double labeled with mitoGFP and TUNEL (red) with lower magnification (E–E″, K–K″, M–M″). Scale bars: 5 µm. MUL1 overexpression is driven by Mef2-Gal4. In wild-type (D), mitochondria have a regular size and shape, and are localized in between myofibrils. In PINK1 mutants (D′), mitochondrial size becomes irregular, and the GFP signal is reduced. Large mitochondrial clumps also appear. PINK1 mutant muscle is TUNEL-positive (E′). (F–F″) Touidine blue staining of muscle. Compared with the wild-type (F), PINK1 mutant muscle shows vacuolation indicating muscle degeneration (F′). These PINK1 mutant phenotypes (D′, E′, F′) are almost completely suppressed by MUL1 overexpression (D″, E″, F″). (Ga–Ga″, Gb–Gb″) EM images of mitochondria in muscle. (Gb–Gb″) Single mitochondrion (outlined with dashed lines) from white boxes in Ga–Ga″. Scale bars: 1 µm (Ga–G″a) 0.5 µm (Gb–G″b). In wild-type (Ga and Gb), mitochondria have compact and organized cristae whereas mitochondria from PINK1 mutants (Ga′, Gb′) are swollen with fragmented cristae, and this is rescued by MUL1 overexpression (Ga″, Gb″). (H) Images of thoraces. Arrows point to thoracic indentations due to muscle degeneration. Compared with WT, PINK1 mutants have thoracic indentation due to muscle degeneration. MUL1 overexpression, but not MUL1 LD overexpression, suppresses PINK1 mutant thoracic indentation. (I) qPCR analysis shows that MUL1 and MUL1 LD mRNA are expressed at similar levels in muscles. The data are shown as the mean ± SEM from three experiments (RNA from ten 5-day-old fly thoraces for each genotype). The statistical analysis was done using One-way ANOVA with Tukey' multiple comparisons test. ns: not statistically significant. MUL1 LD overexpression in the PINK1 mutant background does not suppress the formation of mitochondrial clumps (J″) or TUNEL-positivity (K″). (L–M″) Overexpression of MUL1, but not MUL1 LD, suppresses parkin mutant phenotypes.