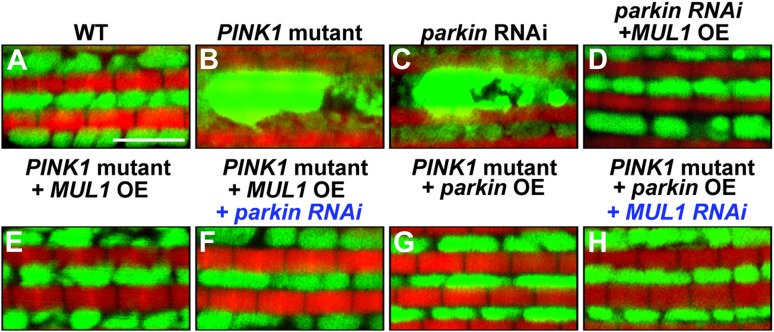
Figure 5. MUL1 acts in parallel to the PINK1/parkin pathway.
(A–H) Images of thoraces of various mutants. Arrows point to thoracic indentations due to muscle degeneration. PINK1 MUL1 and parkin MUL1 double mutants have more severe thoracic indentation compared to either mutant alone. Remarkably, the severe thoracic indentation phenotype in parkin MUL1 double mutants is almost completely suppressed when mfn is also knocked down. (I–P) Mitochondria are labeled using an anti-ATP synthase antibody in the IFM. While PINK1, parkin, and MUL1 mutant show slightly elongated mitochondrial morphology, PINK1 MUL1 and parkin MUL1 double mutants exhibit highly elongated and interconnected mitochondria. These phenotypes can be suppressed by mfn knockdown. Instead of using mitoGFP, we utilized anti-ATPase antibodies that allow better visualization of the enhancement phenotypes seen with double mutants. (Q) Relative ATP levels in whole flies of various mutants (mean ±SEM from three experiments, five 5-day-old flies for each genotype). ** and *** significantly different from wild-type, p<0.01 and p<0.001, respectably (One-way ANOVA with Tukey's multiple comparisons test). # Significantly different from parkin mutant and MUL1 mutant, both p<0.01 (Two-way ANOVA with Tukey's multiple comparisons test). (R) In vivo ubiquitination assay of Mfn. S2 cells were treated with the indicated RNAi, transfected with Flag-Mfn, and treated with proteasome inhibitor MG132. Immunoprecipitations were performed using anti-Flag antibody, and western blots were probed with antibodies against anti-Ubiquitin antibody (P4D1) or anti-Flag antibody. Relative ubiquitination levels compared to control are shown in the lower panel (mean ± SEM). ** and *** Significantly different from control, p<0.01 and p<0.001, respectably (One-way ANOVA with Tukey's multiple comparisons test). # Significantly different from MUL1 RNAi #1 and parkin RNAi, both p<0.01. & Significantly different from MUL1 RNAi #2 and parkin RNAi, p<0.001 and p<0.01, respectably (Two-way ANOVA with Tukey's multiple comparisons test). (S) Western blot analysis of Mfn levels in vivo and quantification (mean ± SEM from three experiments, eight third instar larvae for each genotype). * and ** significantly different from wild-type, p<0.05 and p<0.01, respectably (One-way ANOVA with Tukey's multiple comparisons test). # Significantly different from parkin mutant and MUL1 mutant, both p<0.01. & Significantly different from PINK1 mutant and MUL1 mutant, both p<0.01 (Two-way ANOVA with Tukey's multiple comparisons test).