Abstract
Fluorescence microscopy of GFP-tagged proteins is a fundamental tool in cell biology, but without seeing the structure of the surrounding cellular space, functional information can be lost. Here we present a protocol that preserves GFP and mCherry fluorescence in mammalian cells embedded in resin with electron contrast to reveal cellular ultrastructure. Ultrathin in-resin fluorescence (IRF) sections were imaged simultaneously for fluorescence and electron signals in an integrated light and scanning electron microscope. We show, for the first time, that GFP is stable and active in resin sections in vacuo. We applied our protocol to study the subcellular localisation of diacylglycerol (DAG), a modulator of membrane morphology and membrane dynamics in nuclear envelope assembly. We show that DAG is localised to the nuclear envelope, nucleoplasmic reticulum and curved tips of the Golgi apparatus. With these developments, we demonstrate that integrated imaging is maturing into a powerful tool for accurate molecular localisation to structure.
Keywords: GFP, Mammalian cells, Diacylglycerol, In-resin fluorescence, Correlative light and electron microscopy, Integrated light and electron microscopy
Highlights
-
•
GFP and mCherry fluorescence are preserved in heavy-metal stained mammalian cells embedded in resin
-
•
Fluorophores are stable and intensity is sufficient for detection in ultrathin sections
-
•
Overlay of separate LM and EM images from the same ultrathin section improves CLEM protein localisation precision
-
•
GFP is stable and active in the vacuum of an integrated light and scanning EM
-
•
Integrated light and electron microscopy shows new subcellular locations of the lipid diacylglycerol
1. Introduction
Correlative Light and Electron Microscopy (CLEM) uses separate light and electron microscopes to localise cloned fluorophores within the structural reference space of the cell [1,2]. Fluorophores are first imaged using light microscopy, followed by an extended sample preparation procedure, after which ultrathin sections of resin-embedded cells are cut and viewed in the electron microscope (EM). The use of separate imaging modalities makes the accurate localisation of a fluorescent signal to specific cellular structures difficult, particularly as the axial resolution of a standard fluorescence microscope is approximately one order of magnitude lower than that of an ultrathin resin section imaged in the EM.
Recent advances in imaging systems have led to dual modality instruments for integrated light and electron microscopy (ILEM). The integrated laser and electron microscope (iLEM) enables dual detection of signals by embedding a laser-scanning fluorescence microscope into a transmission electron microscope (TEM) [3–5]. More recently, a widefield epifluorescence microscope has been integrated within a scanning electron microscope (SEM) for ‘simultaneous CLEM׳ (sCLEM) [6,7]. These integrated imaging systems require maintenance of dual fluorescence and electron signals within the same biological sample, as well as stability and activity of the fluorophore within the microscope vacuum.
Protocols have been reported that preserve fluorescence in-resin for serial light microscopy [8], but post-embedding staining is required for subsequent electron imaging [9] and so the samples are not suitable for imaging in integrated microscopes. An approach that delivers dual fluorescence and electron signals in the same sample for integrated microscopy is post-embedding immunolabelling of ultrathin sections (resin or Tokuyasu) with antibodies conjugated to relatively photostable fluorophores (such as TRITC, Alexa dyes and DAPI) for iLEM [4,10–13] and sCLEM [6,7]. The alternative is to produce an ultrathin section of a resin-embedded cell that contains a detectable signal from expressed fluorescent proteins alongside sufficient staining of the cell membranes to introduce electron contrast for structural interpretation. This is a significant challenge, as many of the EM processing steps are incompatible with fluorescence preservation, including dehydration and heavy metal staining. Protocols have been described for in-resin fluorescence (IRF) of various fluorophores in samples ranging from viruses and yeast [14,15] to Caenorhabditis elegans [16,17], zebrafish [18,19], Drosophila embryos [20] and plants [21]. These techniques have great potential for the accurate correlation of molecules to structure in cell biology. However, to-date, imaging of resin-embedded GFP fluorescence in situ in an integrated microscope has not been reported. Here, we address this ‘missing link’ by developing a new IRF protocol for mammalian cells that delivers stable and active fluorescence in resin sections in the vacuum of an integrated light and scanning electron microscope (ILSEM).
We have applied this technique to a technically challenging study, investigating the distribution of the lipid diacylglycerol (DAG) within cellular membranes. We have previously demonstrated that DAG has a dual role as a modulator of membrane dynamics and as a second messenger in mammalian and echinoderm cells [22–24]. We have also used confocal microscopy and CLEM as tools to analyse mammalian cells in which DAG was acutely and specifically depleted from the nuclear envelope and endoplasmic reticulum, which led to disruption of nuclear envelope reformation at mitosis [24]. Analysis of the distribution of DAG within the nuclear envelope is therefore expected to contribute to an understanding of its function in this process. However, subcellular lipid localisation is a challenging task, both in terms of probes and imaging.
Immunolabelling of DAG is problematic because it does not have a head-group, and so specific antibodies have not been generated. In the absence of antibodies, lipids can be localised by labelling with purified recombinant phospholipid recognition domains, or by transfection of fluorescently-labelled phospholipid domain probes [25–27]. Here, we use a GFP-C1a-C1b probe from PKCε [24], which must be transfected into cells, as the recombinant version of the probe is relatively unstable. We performed post-embedding CLEM on IRF sections using separate light and electron microscopes, resulting in improved localisation accuracy when compared to pre-embedding CLEM performed on whole cells. The fluorescent signal for DAG localised to the nuclear envelope, nucleoplasmic reticulum subdomains, and the Golgi apparatus, where it was possible to detect a higher concentration in the curved tips of individual Golgi cisternae (indicated by higher intensity fluorescent signal). Finally, we confirmed the subcellular localisation of DAG in serial ultrathin sections in an integrated light and scanning electron microscope. This is the first report of GFP fluorescence in resin-embedded biological samples in vacuo correlated to subcellular structure, and as such, introduces a powerful new imaging tool for structure/ function studies.
2. Results
2.1. Localisation of DAG to cellular membranes by CLEM (pre-embedding LM)
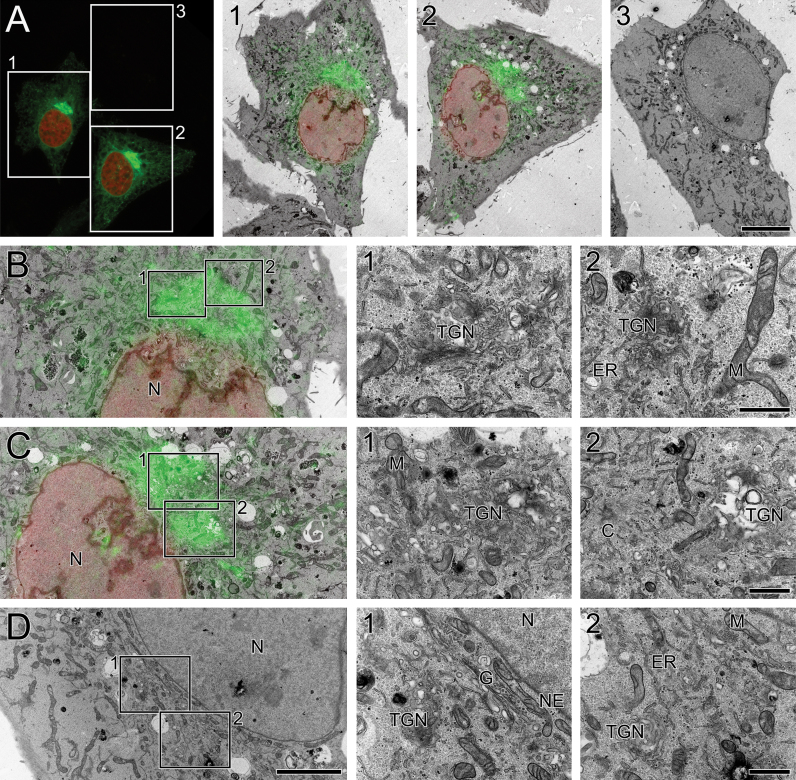
HeLa cells were transfected with GFP-C1 and mCherry-H2B and imaged using confocal laser scanning microscopy (Fig. 1A) with an axial resolution of 0.7 μm. Cells were then processed for CLEM, the cells of interest were relocated laterally within the block and serial sections of 70 nm were collected and imaged in the TEM. Confocal and TEM images were overlaid to find the closest match of fluorescence signal to structure (Fig. 1A, 1 and 2), bearing in mind that each confocal image corresponds to a series of 10 EM images through the z-plane. The overlay accuracy was low due to the difference in axial resolution between LM and EM, shrinkage during sample processing for EM, and compression during sectioning. As expected, the mCherry-H2B signal localised to the nucleus (Fig. 1B,C). GFP-C1 was found at the periphery of the nucleus, indicating localisation of DAG to the NE as previously reported [24]. GFP-C1 was also concentrated in the perinuclear region, which was rich in trans-Golgi network (TGN) membranes as revealed by EM (Fig. 1B,C, 1 and 2). However, due to the intensity of the fluorescence in this region, emanating from a volume ten times that of the EM section plane, it was difficult to unambiguously assign the GFP signal to TGN membranes. Similarly, peripheral distribution of GFP-C1 suggested localisation of DAG to the endoplasmic reticulum (ER), but the alignment of fluorescent signal to structure was poor. To improve the axial resolution of the fluorescence signal and increase overlay accuracy and protein localisation precision, we developed a protocol for preservation of GFP and mCherry signals in mammalian cells after EM sample preparation.
Fig. 1.
CLEM of GFP-C1 and mCherry-H2B in HeLa cells using pre-embedding light microscopy. (A) Maximum intensity projection of a confocal microscopy image stack showing two cells expressing GFP-C1 and mCherry-H2B and an adjacent cell which is not transfected. Electron micrographs of the boxed regions are shown in panels 1–3. For each micrograph, the closest approximate slice from the confocal image stack has been used for overlaying the fluorescent signal onto the electron micrograph. (B, C) Electron micrographs showing localisation of the fluorescent signal to specific structures within the cell; mCherry-H2B is clearly localised to the nucleus, whereas GFP-C1 expression is strongly associated with the perinuclear region. GFP-C1 is less clearly localised to the nuclear envelope and endoplasmic reticulum. (D) Electron micrographs showing a similar perinuclear region within the adjacent cell, which is not transfected. C: centriole, ER: endoplasmic reticulum, G: Golgi, M: mitochondrion, N: nucleus, NE: nuclear envelope, TGN: trans-Golgi network. Scale bars—A: inset 1/2/3: 10 μm, B–D: 5 μm (inset 1/2: 1 μm).
2.2. High pressure freezing and quick freeze substitution for IRF of GFP in mammalian cells
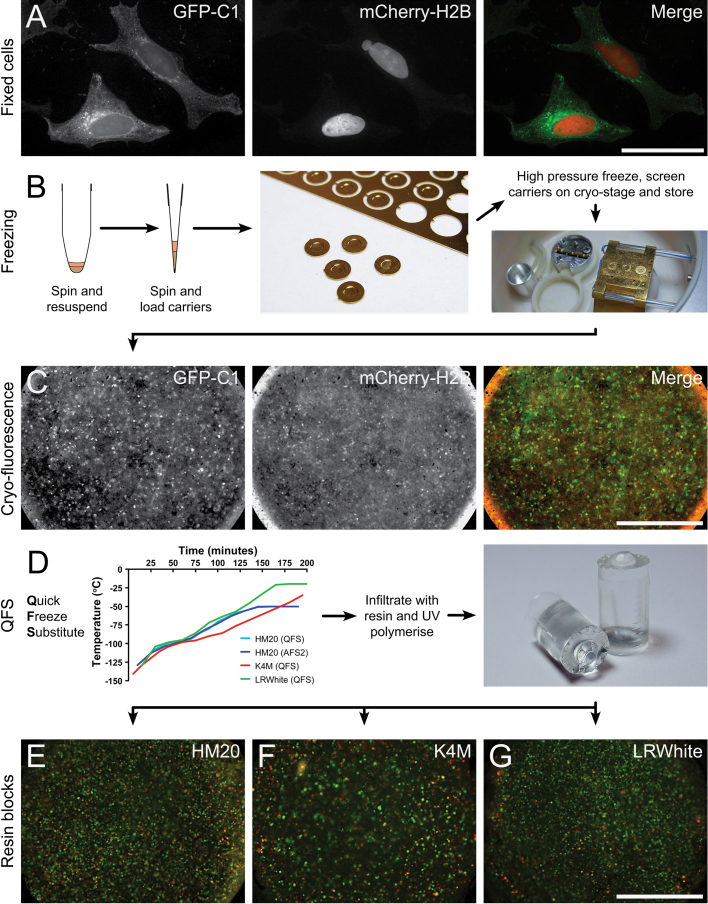
HeLa cells were grown in 10 cm tissue culture dishes and transfected with GFP-C1 and mCherry-H2B constructs (Fig. 2A). The cells were trypsinised, pelleted, loaded into planchettes and high pressure frozen (Fig. 2B), thus avoiding the fluorophore quenching that can occur during chemical fixation with aldehydes. Cells suspended in vitreous ice were screened for GFP and mCherry fluorescence at −196 °C using a cryo-fluorescence stage (Fig. 2B,C), prior to freeze substitution (FS).
Fig. 2.
Preservation of fluorescence in resin through high pressure freezing (HPF) and quick freeze substitution (QFS). (A) Images from a confocal image stack of fixed cells grown on coverslips. The cells are expressing both GFP-C1 and mCherry-H2B. (B) Schematic illustrating the method used to prepare cells for freezing. Left: cells are resuspended in an equal volume of media containing 10% BSA, and spun again in a blocked tip. Centre: the tip blockage is cut away and the cells are pipetted into membrane carriers. Right: the carriers are high pressure frozen and screened on the cryo-correlative stage before storage. (C) Images showing that fluorescence is preserved after high pressure freezing. Screening of carriers in this way not only confirms successful preservation of fluorescence, but also confirms presence of a sufficient number of cells in the carrier for continuation of processing. (D) Carriers are processed for quick freeze substitution (QFS). Left: graph showing the typical temperature profiles recorded during QFS using the polystyrene box method, and replicated in the AFS2. Variations in the temperature profiles recorded reflect differences in room conditions on the day of processing. Middle: after substitution, samples are infiltrated with resin and polymerised under UV light. Right: image showing two polymerised HM20 blocks, the tips of which contain discs of cells from the membrane carrier. (E–G) Merged images showing preserved GFP-C1 and mCherry-H2B fluorescence in resin-embedded cells directly imaged from the face of the resin block (HM20, K4M, and LR White, respectively). Scale bars—A: 10 μm, C, D: 500 μm.
Cells were initially subjected to an FS protocol reported to preserve GFP in zebrafish [19], but this resulted in quenching of both GFP and mCherry fluorescence. We postulated that the loss of signal was due to prolonged exposure to solvents, leading to removal of the hydration shell, which can affect GFP dynamics [28]. Dehydration effects may be more severe in cells than in zebrafish or worms, as cells are at least an order of magnitude smaller. Therefore, we minimised FS time using a Quick FS protocol developed by McDonald & Webb [29] (Fig. 2D) which resulted in GFP and mCherry fluorescence preservation in HM20, K4M and LR White resins (Fig. 2E–G). The FS was carried out in a polystyrene box, and then reproduced in an automated freeze substitution unit. Fluorescence was preserved in both, suggesting that speed is key to the success of the technique. Critically, we found that fluorophores within the resin blocks were remarkably stable. In optimally embedded samples, GFP fluorescence can be imaged in resin fifteen months (or more) after polymerisation. The protocol was repeated using COS-7 cells expressing the same fluorescent constructs, which also resulted in successful GFP and mCherry preservation (data not shown).
2.3. Light microscopy of fluorophores in ultrathin resin sections
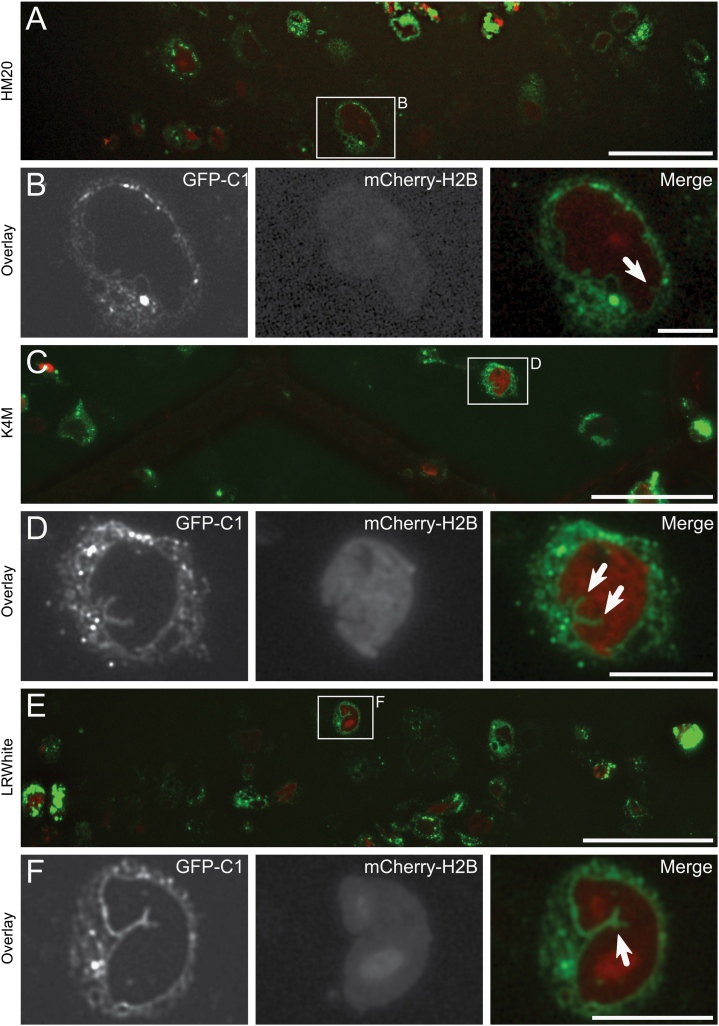
Ultrathin sections (70 nm) were cut from resin-embedded cells expressing GFP-C1 and mCherry-H2B embedded in HM20 (Fig. 3A,B), K4M (Fig. 3C,D) and LR White (Fig. 3E,F). The sections were imaged using a standard widefield epifluorescence microscope. Though signal intensity was low, requiring exposure times of around 6–8 s with our microscope and camera set-up (using the 100✕ objective with dry imaging conditions; 3–4 s using the 40× objective with wet imaging conditions), both GFP and mCherry could be detected in all three resins.
Fig. 3.
Light microscopy of GFP-C1 and mCherry-H2B in 70 nm resin sections using a widefield epifluorescence light microscope. (A) Overview of a 70 nm thick section through cells embedded in HM20, collected on a finder grid and imaged in phosphate buffered glycerol. A wide variety in expression level of both constructs can be seen. (B) Enlarged detail of an individual cell boxed in (A) showing mCherry-H2B within the nucleus, and GFP-C1 associated with the nuclear envelope and structures within the cytoplasm. (C, D) As above, for cells embedded in K4M. (E, F) As above, for cells embedded in LR White. White arrows indicate nucleoplasmic reticulum. Scale bars—A, C, E: 50 μm, B, D, F: 10 μm.
When comparing imaging conditions using sections from optimally embedded cells, we noted that the signal intensity was much stronger when grids were mounted in buffered glycerol than seen in dry imaging conditions. This finding is similar to Karreman et al. [13] who showed that Alexas 488 and 532 exhibited brighter intensities when imaged in water than in air. However, it is important to note that both GFP and mCherry could be imaged in dry conditions, which is an essential starting point for sections to be imaged inside the vacuum of an integrated light and electron microscope. A reproducible gradient of electron contrast was noted through the depth of the cell layer, presumably a result of a heavy metal gradient through the volume (data not shown). Though not quantified, we also noted that levels of preserved fluorescence in some samples exhibited an opposing gradient, and that preservation of fluorescence was often better where electron contrast was poorest.
2.4. Localisation of DAG to cellular membranes by CLEM (post-embedding LM)
GFP-C1 distribution was analysed in ultrathin resin sections using widefield epifluorescence microscopy. DAG was localised to the nuclear envelope, as well as to structures invaginating into the nucleus (arrows) and punctae of different sizes in the cytosol (Fig. 3B,D,F). Sections that had been imaged in the fluorescence microscope were then transferred directly to the transmission electron microscope without any additional heavy-metal staining (Fig. 4). Cells of interest were relocated using the finder pattern on the grids, or simply by cell position in the section. Following EM image acquisition the fluorescence and electron signals were overlaid. Unlike CLEM protocols using pre-embedding light microscopy (Fig. 1), only small linear adjustments were required for an accurate overlay. Furthermore, the axial resolution of fluorescence and electron signals was the same, thus improving protein localisation precision. HM20 (Fig. 4A,B) and LR White gave the best contrast and ultrastructure, whereas K4M resin tended to infiltrate and section poorly. An example of overlays from two consecutive HM20 sections is shown (Fig. 4A,B), demonstrating the potential for serial sectioning and imaging to deliver 3D LM/ EM overlays with 70 nm axial resolution in both imaging modalities.
Fig. 4.
CLEM of GFP-C1 and mCherry-H2B in HeLa cells using post-embedding light microscopy. GFP-C1 fluorescent signals overlaid onto electron micrographs of the corresponding region using the IRF method. (A) Image of GFP-C1 signal alone imaged under dry conditions, and overlaid directly onto the matching electron micrograph, for a cell embedded in HM20. The fluorescent signal corresponds to the nuclear envelope, and other structures within the cytoplasm (higher magnification micrographs of subcellular detail are shown in Fig. 5). (B) An adjacent section through the cell shown in A. (C–E) Increased precision of GFP-C1 localisation to Golgi stacks (G) and the NE. In (C), black arrows indicate localisation of fluorescent signal to the highly curved tips of Golgi cisternae. An enlarged view from an adjacent section is shown in (D). Localisation to the NE is also evident. In (E), white arrows indicate localisation of fluorescent signal to the nucleoplasmic reticulum. (F, G) Localisation of GFP-C1 to endoplasmic reticulum and to membrane stacks (black arrows) in cells expressing low (F) and high (G) levels of the GFP-C1 construct. ER: endoplasmic reticulum, G: Golgi, M: mitochondrion, N: nucleus, NE: nuclear envelope. Scale bars—A, B: 5 μm, C, D: 500 nm, E-G: 1 μm.
Higher magnification analysis of IRF overlays revealed improved localisation precision of DAG to cellular membranes using IRF (Fig. 4C–G) compared to CLEM protocols using pre-embedding light microscopy (Fig. 1). We found that DAG was particularly concentrated at the tips of Golgi cisternae (black arrows, Fig. 4C,D). DAG was also localised to the NE (Fig. 4C–E), and the intranuclear localisation seen in fluorescence microscopy of the IRF sections was associated with invaginations of the nuclear envelope, also known as nucleoplasmic reticulum (Fig. 4E, white arrows). DAG was also localised to the ER, and could be seen in cells expressing both low (Fig. 4F) and high (Fig. 4G) levels of the GFP-C1 construct.
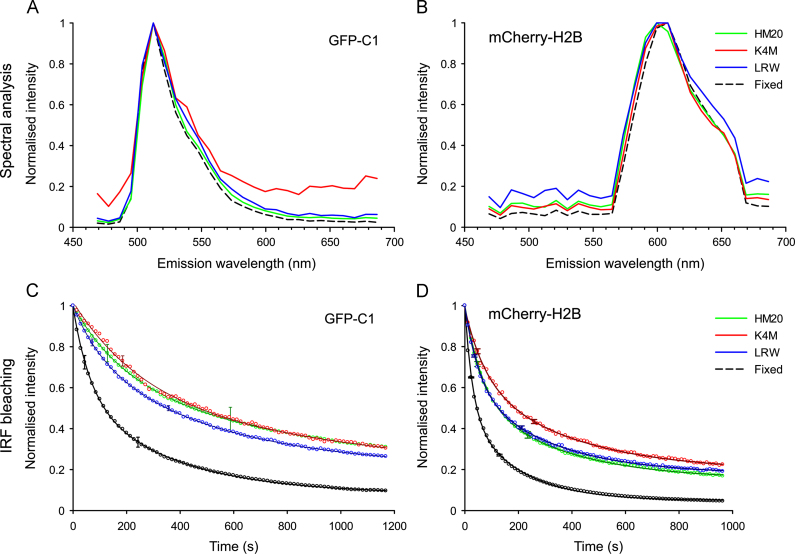
2.5. Validation of fluorophore signals by spectral analysis
Spectral analysis was used to confirm that fluorescent signals resulted from the cloned fluorophores and were not a result of artefacts such as holes and tears in the resin caused by poor embedding, resin autofluorescence or anomalous signals from stained structures in the cells. Emission profiles for both GFP and mCherry were identical in the fixed and mounted whole cells as well as in all three resins (Fig. 5A,B). Some background autofluorescence was noted, particularly for K4M in the GFP emission profile and LR White in the mCherry emission profile. However, this was not strong enough to interfere with the true GFP or mCherry signals, and reduced over time from embedding relative to the cloned fluorophores.
Fig. 5.
Validation of GFP and mCherry signals in IRF sections by spectral profiling. (A, B) Spectral profile of GFP (A) and mCherry (B) in fixed and mounted cells, and in each of the three resins, showing normalised intensity across the lambda stack. No significant shift in emission profiles was noted aside from changes in background noise characteristics. (C, D) The bleaching rates of resin embedded GFP (C) and mCherry (D) were compared to fixed and mounted cells; the fixed cells showed much higher rates of fluorophore bleaching over time. Error bars indicate standard deviation at the nearest point to the t½ value.
We further characterised the in-resin fluorescence and found that the intensity of fluorophores in the resin sections was significantly reduced compared to fixed and mounted whole cells. This can be explained in part by fewer fluorochromes being available in the 70 nm section than in the whole cell, in addition to expected losses through sample processing. However, the preserved fluorescence within the resin sections appeared to be significantly more photostable than in the fixed and mounted whole cells (Fig. 5C,D). Indeed, analysis showed that fluorophores in the resin sections were 2–5 times more resistant to photobleaching, with a small proportion that proved very difficult to bleach. This was in stark contrast to the fixed cells that rapidly bleached to near-background. Due to the complex nature of the bleaching curves, simple single or double exponential fits were not well suited, as may be expected [30]. However, for relative comparisons a double exponential was used in this instance. The relative t½ under our imaging conditions were as follows: fixed and mounted samples exhibited GFP t½ of 45±1 s (43%) and 244±5 s (49%) for the fast and slower fractions, respectively. This compared to HM20 GFP t½ of 129±7 s (33%) and 587±75 s (48%) and LR White GFP t½ of 76±4 s (22%) and 350±13 s (57%). mCherry also showed a similar trend with fixed and mounted samples exhibiting a t½ of 20.3±0.5 s (52%) and 132±3 s (43%), HM20 mCherry a t½ of 44±1 s (43%) and 230±8 s (42%), K4M mCherry a t½ of 50±4 s (33%) and 255±17 s (48%) and LR White mCherry a t½ of 36±1 s (38%) and 208±6 s (44%).
2.6. Localisation of DAG to cellular membranes by ILEM
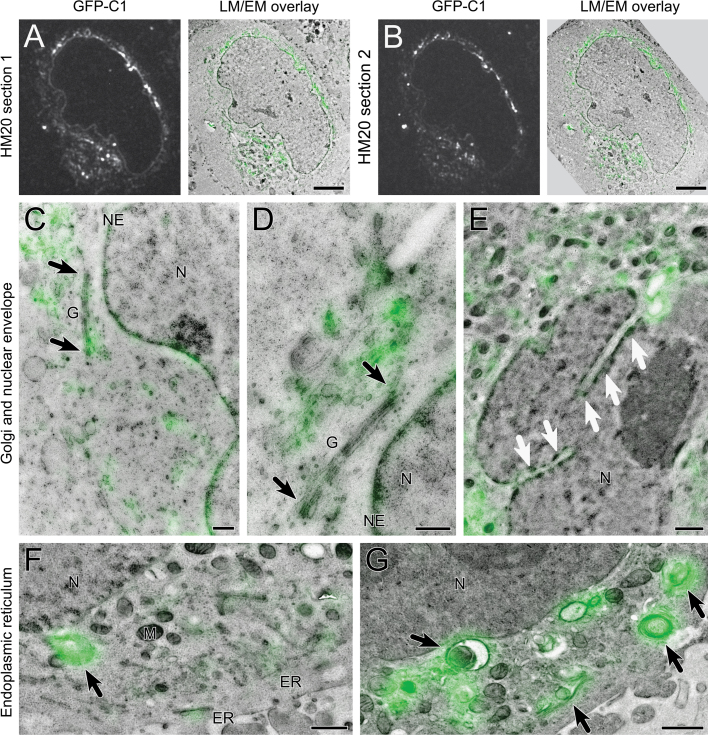
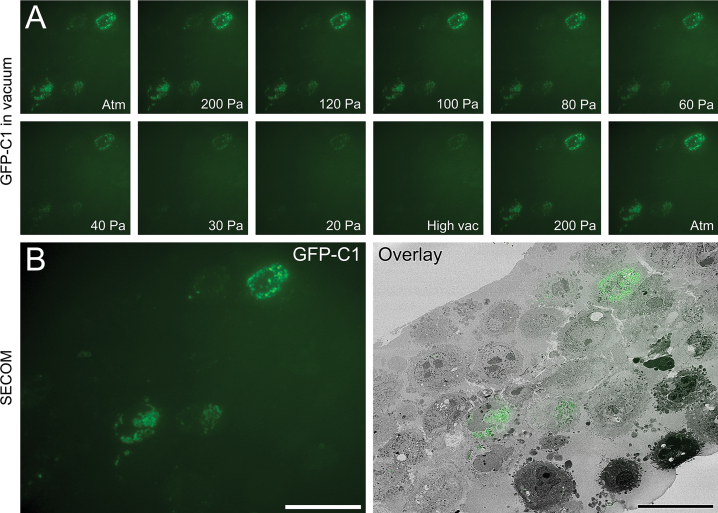
Fluorescence and electron images of 200 nm resin sections were collected in a SECOM ILSEM (DELMIC; Fig. 6) equipped with a widefield epifluorescence light microscope. GFP in IRF sections was active within the vacuum of the electron microscope. This enabled preliminary imaging experiments to test the optimal in vacuo imaging conditions. Interestingly, the GFP signal varied with vacuum pressure, with a drop in intensity as the pressure decreased towards high vacuum conditions (Fig. 6A). This was not due to photobleaching from repeated imaging, as increasing the chamber pressure to 200 Pa and then to atmospheric pressure (Atm) resulted in recovery of the signal. In fact, GFP was stable and resistant to photobleaching over a period of at least 10 min whilst the image series was collected.
Fig. 6.
GFP in IRF sections is stable and active at atmospheric pressure and in vacuum. (A) Sequential images depicting the effect of vacuum pressure on the fluorescent signal recorded from a 200 nm IRF section, beginning at atmospheric pressure (Atm), proceeding stepwise to high vacuum, and returning to atmospheric pressure. (B) Overlaid fluorescent signal for GFP-C1, recorded at 200 Pa, onto the matching scanning electron micrograph recorded in high vacuum. Scale bars—25 μm.
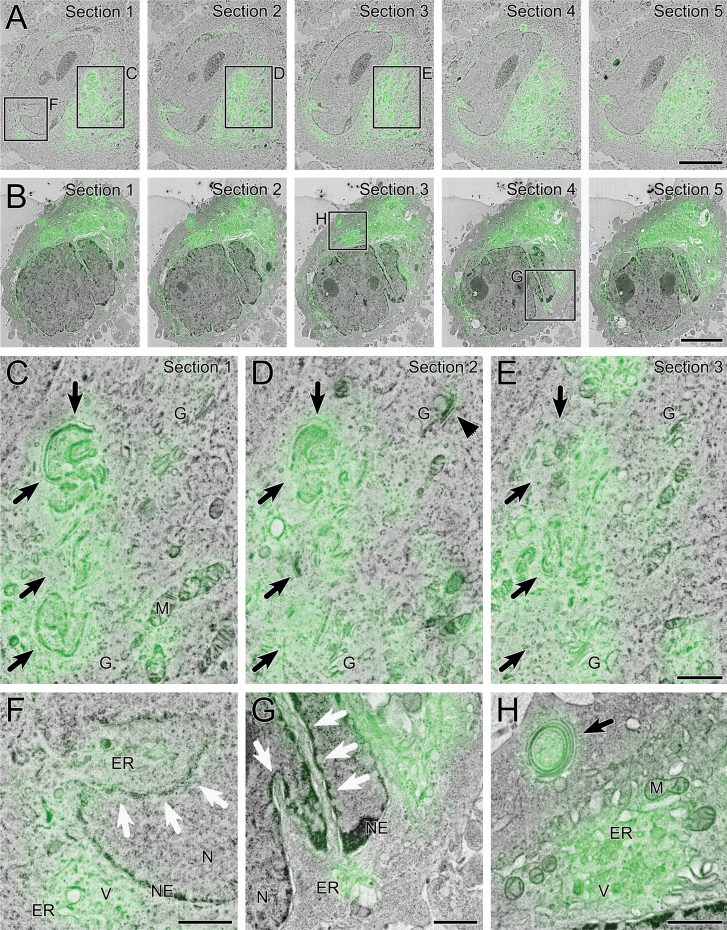
Electron imaging of IRF sections was carried out in situ immediately after fluorescence imaging (Fig. 6B; Fig. 7). The fluorescent signal could be followed in the same cell over multiple serial sections (Fig. 7A,B). DAG fluorescence was localised to membranous structures within the cytoplasm (black arrows, Fig. 7C–E), Golgi cisternae (G and black arrowhead; Fig. 7D), nucleoplasmic reticulum (white arrows, Fig. 7F,G), endoplasmic reticulum (Fig. 7F–H), and patches of complex vesicular structures (Fig. 7H).
Fig. 7.
ILEM of GFP-C1 in HeLa cells. GFP-C1 fluorescent signals overlaid onto electron micrographs of the corresponding region using the IRF method, acquired using an ILSEM. (A, B) Images from serial HM20 sections through cells expressing comparatively low (A) and high (B) levels of the GFP-C1 construct. The fluorescent signal corresponds to the nuclear envelope and nucleoplasmic reticulum, and other membranous structures within the cytoplasm including Golgi stacks and endoplasmic reticulum. (C–E) Boxed detail from serial Sections 1–3 in A. The fluorescent signal corresponds to curved membrane stacks (black arrows) and curved tips of Golgi cisternae (G and black arrowhead). (F) Boxed detail from Section 1 in A showing fluorescent signal at the nucleoplasmic reticulum (white arrows). The invagination contains GFP-C1 positive endoplasmic reticulum, which is also visible outside the nucleus with other small vesicular structures. (G) Boxed detail from Section 4 in B showing fluorescent signal associated with the nucleoplasmic reticulum (white arrows), and segments of endoplasmic reticulum surrounding the nucleus. (H) Boxed detail from Section 3 in B showing GFP-C1 signal corresponding directly to a coiled membrane stack (black arrow). Intense fluorescent signal correlates to an area of cytoplasm containing complex membranous structures including segments of endoplasmic reticulum and vesicles. ER: endoplasmic reticulum; G: Golgi; M: mitochondria; N: nucleus; NE: nuclear envelope; V: vesicles. Scale bars—5 μm (A, B), 1 μm (E-H).
These findings demonstrate the potential of the IRF protocol combined with integrated microscopy to deliver fast, seamless, dual modality imaging for accurate protein localisation in cell biology.
3. Discussion
GFP expression can be contextualised using CLEM to reveal the surrounding structural reference space of the cell. Correlative microscopy using two separate imaging systems is now developing into integrated microscopy, allowing light and electron signals to be detected in situ without further specimen manipulation steps. Integrated imaging has the potential to speed up the process, and significantly improves image overlay accuracy and protein localisation precision. However, protocol development for the preparation of samples with dual signals has not kept pace with technological advances.
Fluorophores have been preserved in resin-embedded cultured cells without en bloc heavy-metal staining [9], and in heavy-metal stained and resin-embedded yeast, zebrafish, C. elegans, D. melanogaster and plants. We now extend the available protocols to include preservation of fluorescence from GFP- and mCherry-tagged proteins in cultured mammalian cells with electron contrast for structural analysis. Our protocol includes low concentrations of water and uranyl acetate in the substitution medium and is extremely short. In fact, we believe that the inclusion of QFS [29] within the IRF protocol may be essential to produce cellular samples with the necessary characteristics for ILEM: fluorophores that are stable during imaging and long-term storage, and robust enough to be detected in ultrathin sections (70–200 nm) as opposed to the thicker sections (>300 nm) required in other studies [9,21].
Imaging IRF sections on separate light and electron microscopes delivers better overlay accuracy than CLEM performed with pre-embedding light microscopy of whole cells due to the matching axial resolution of the images. However, our ultimate aim was to image the IRF sections in situ in the ILSEM for fast, seamless imaging of dual fluorescence and electron signals with high precision [6,7]. For the first time, we demonstrate GFP fluorescence in resin-embedded biological samples in vacuo. The GFP signal was stable, and sufficiently intense to detect a range of expression levels of the GFP-C1 construct in cells. Switching from fluorescence to electron imaging is simple, with the potential for overlay accuracies in the order of 10 nm [7] due to the precise alignment of the light and electron beams.
We demonstrate the application of this new integrated imaging workflow by extending our recent study on the involvement of diacylglycerol in the re-formation of the nuclear envelope at mitosis. We show for the first time that intranuclear DAG is associated with nucleoplasmic reticulum membranes, indicating a role in localised membrane morphology, as well as defining an unprecedented site of localisation within the nucleus. This also raises the question of whether all lipids that are apparently located in the interior of the nucleus remain associated with membranes. Evidence for interior membranes unconnected to the NE is scant [31] but has been reported [32], which further complicates assignment of interior phospholipids to non-membranous origins. Future work will involve manipulation of DAG levels in interphase cells to investigate functional roles of lipids in the nucleoplasmic reticulum.
The increased accuracy of IRF overlays also indicates an asymmetric distribution of DAG in the Golgi apparatus, where it is more concentrated at the curved tips of cisternae. This supports the idea that highly curved membranous regions require the presence of spontaneously negatively curved lipids such as DAG, and indicates an additional role for DAG in organelle shaping [24]. However, the improvement in overlay accuracy is difficult to quantify because the GFP fluorescence is distributed throughout organelles rather than emanating from a ‘point’ source such as a virus particle or a clathrin-coated vesicle [14,15]. In addition, though the axial resolution of the images far exceeds the diffraction limit of light due to the physical section thickness [9,17], at present we are still working with diffraction-limited lateral resolution. Improvements could be made to the fluorescence images by integrating a super-resolution light microscope within the SEM, a logical extension of the STED imaging of IRF sections reported by Watanabe et al. [17].
The ability to directly detect GFP using ILEM puts a new and powerful tool into the hands of cell biologists. Ultrastructural protein localisation is no longer dependent on the availability and affinity of primary antibodies. There is no longer a requirement for permeabilisation of membranes to allow access of antibodies into the sample, which invariably disrupts the ultrastructure, so that integrated imaging is scalable for protein localisation within tissues and model organisms. Work is now underway to automate this process in the next generation of serial imaging systems for fast, fully aligned 3D ILEM using serial imaging techniques like array tomography [33], Serial Block Face SEM and Focused Ion Beam SEM [34].
Finally, the combination of IRF and ILEM could lead to ‘multi-colour’ electron microscopy. At present, it is possible to immunolabel with several antibodies using different immunogold particle sizes, though species specificity of the primary antibody is usually a limitation. Using IRF and ILEM, the main limitations on the number of proteins localised will be the number of fluorescent probes within the sample and the range of wavelengths excited and detected by the integrated light microscope. Thus, many pre-existing cell lines and model organisms expressing fluorescent proteins could now be accessible to high precision protein localisation studies.
4. Materials and methods
4.1. Cell culture, constructs and transfection
Human cervical cancer epithelial HeLa cells were obtained from the American Type Culture Collection (ATCC, CCL-2). Cells were maintained in DMEM supplemented with 10% foetal bovine serum in 10 or 15 cm tissue culture dishes. PKCε and mCherry-H2B were kindly provided by Peter J. Parker (London Research Institute, UK).
EGFP-PKCε was made by removing an internal BamHI site using the oligos:
Sense–gccccacaagttcggcatccacaactacaaggtccccacg
Antisense–cgtggggaccttgtagttgtggatgccgaacttgtggggc
PKCεC1aC1b was PCR amplified using the oligos:
Sense–agggatccatgcacaacttcatggccacctacttgcggcaac
Antisense–ttgccgcaagtaggtggccatgaagttgtgcatggatccct
Cells were transfected with 0.5 μg DNA (for 3.5 cm MatTek dishes) of each construct using Lipofectamine LTX and PLUS reagent (Invitrogen, Life Technologies Ltd, Paisley) in OPTIMEM medium (Gibco, Life Technologies Ltd, Paisley) as recommended by the manufacturer. The transfection mix was added to the cells in antibiotic-free medium. Microscopy and/or fixation was performed 18–24 h after transfection.
4.2. CLEM with pre-embedding light microscopy
Cells were grown on gridded glass coverslips in MatTek dishes (MatTek Corporation, Ashland, MA, USA), fixed in 4% paraformaldehyde in 0.1 M phosphate buffer (PB), pH 7.4, and mounted in Mowiol/DABCO prior to imaging for brightfield and fluorescence signals on an inverted LSM 710 confocal microscope with a 63×/1.4 objective (Zeiss, Cambridge). Secondary fixation was performed in 1.5% glutaraldehyde/2% paraformaldehyde in 0.1 M PB for 30 mins. After fixation, coverslips were carefully removed from the MatTek dishes and washed several times in 0.1 M PB. The cells were post-fixed in 1.5% potassium ferricyanide/1% osmium tetroxide for 1 h, before rinsing in PB, and incubating in 1% tannic acid in 0.05 M PB (w/v) for 45 min to enhance membrane contrast. After a brief rinse in 1% sodium sulphate in 0.05 M PB (w/v), the coverslips were washed twice in distilled water, dehydrated through an ascending series of ethanol and propylene oxide prior to infiltration with epoxy resin and polymerisation overnight at 60 °C. The coverslips were removed from the resin blocks by plunging briefly into liquid nitrogen. The cells of interest were identified by correlating the grid and cell pattern on the surface of the block with previously acquired brightfield images. The area of interest was cut from the block and further trimmed by hand using a single-edged razor blade to form a small trapezoid blockface. Using a diamond knife, serial ultrathin sections of 70 nm thickness were cut through the depth of the cells of interest and collected on 1.5% formvar-coated single slot grids. The sections were counterstained with lead citrate to further enhance contrast prior to electron imaging.
4.3. High pressure freezing
A schematic representation of the preparation method used is shown in Fig. 2. For high pressure freezing, the cells were spun in an Eppendorf tube to form a pellet, resuspended in an equal volume of media containing 10% BSA, and maintained at 37 °C. For loading into membrane carriers, a volume of cells was spun down in a blocked 200 μl pipette tip. After cutting away the end of the tip, the cells were pipetted into membrane carriers, loaded into the EMPACT2 high pressure freezer using the rapid transfer system (Leica Microsystems, Vienna) and high pressure frozen. Carriers containing frozen cells were stored under liquid nitrogen.
4.4. Cryo-fluorescence
To monitor levels of fluorescence after high pressure freezing, some carriers from each batch of frozen cells were screened using a cryo-correlative stage (CMS-196, Linkam Scientific Instruments, Chilworth) mounted on a standard epifluorescence microscope (Axio Scope, Zeiss, Cambridge). Carriers containing frozen cells were transferred to the stage under liquid nitrogen and placed directly on the viewing platform for imaging at −196 °C. Processing of screened and non-screened cells in parallel showed a negligible effect of screening on fluorescence preservation and ultrastructural preservation (data not shown).
4.5. Freeze substitution and quick freeze substitution
Quick freeze substitution (QFS) was performed using a modified version of the method described by McDonald and Webb [29]. Tubes containing substitution media (5% H2O in acetone, with either 0.1% or 0.2% uranyl acetate diluted from 20% stock in methanol) were placed in a metal block cooled to −196 °C within a polystyrene box and frozen. Carriers containing high pressure frozen cells were transferred to the tubes containing frozen substitution media under liquid nitrogen. After carrier transfer, the liquid nitrogen in the tube was poured away and the tubes were capped and returned to the cooled block. To initiate substitution, the liquid nitrogen was removed from the box and replaced with dry ice. The temperature of the cooled block was monitored using a temperature probe mounted within a tube in the block, and the dry ice was removed once the block reached −85 °C. At this point, the block was turned on its side within the box and placed on a rotary shaker at 100 rpm.
For embedding in HM20 resin, the tubes were transferred to an automated freeze substitution unit (AFS2; Leica Microsystems, Vienna) once the block reached −50 °C and held at this temperature until the total substitution run-time reached 3 h. For embedding in K4M resin, the transfer and holding temperature was −35 °C, and for LR White, −20 °C. Where the temperature was not reached by 3 h, the tubes were transferred as soon as the holding temperature was reached. The carriers were then gently transferred to moulds filled with 100% acetone and incubated for 15 min. A further 3×15 min washes with 100% acetone were carried out prior to resin infiltration carried out over 3 h through 20/40/60/80/100% dilutions of resin. After incubating overnight in 100% resin, a further 4 changes of fresh resin were carried out the following day, prior to polymerisation under 360 nm UV light over 48 h, and warming to room temperature.
Graphs illustrating the typical temperature profile recorded during QFS are shown in Fig. 2. QFS temperature profiles tended to vary with fluctuations in room environment, therefore we tested a more strictly controlled QFS performed in the automated FS unit. The AFS2 was programmed to closely mimic the typical temperature profile recorded during freeze substitution. This method also produced blocks in which fluorescence was preserved, and the quality of resin infiltration and embedding was comparable to that of the polystyrene box QFS method (data not shown).
Polymerised blocks were trimmed from the moulds and stored at room temperature in the dark. Prior to sectioning, the membrane carriers were carefully trimmed away by hand, and the blocks cut and trimmed perpendicular to the cell layer to allow examination of the full depth of the layer. Ultrathin sections of 70 nm and 200 nm thicknesses were cut using a UCT ultramicrotome (Leica Microsystems, Vienna), collected on 1.5% formvar-coated finder grids and slot grids, and stored at room temperature in the dark.
4.6. Fluorescence imaging of IRF sections
Ultrathin sections were imaged for fluorescent signal in two ways, depending on the quality of resin infiltration and embedding. Where embedding was less than optimal, it proved necessary to image sections hydrated to eliminate reflections from holes and tears in the sections. For this, grids were mounted between a glass slide and coverslip using phosphate buffered glycerol, pH 8.6. However, in optimal conditions this step was unnecessary, and the fluorescent signal could be directly collected from grids placed upon glass slides. Images were collected with the standard widefield epifluorescence microscope using 40× or 100× air objectives (EC Plan-Neofluar 40×/0.75; EC Epiplan-Neofluar 100×/0.75) and an MRm CCD camera (Zeiss, Cambridge).
4.7. Electron imaging of IRF sections
Electron microscopic examination and digital image acquisition was conducted on a Tecnai G2 Spirit BioTWIN (FEI Company, Eindhoven) with an Orius CCD camera (Gatan Inc., Pleasanton). Cells of interest expressing GFP-C1 and mCherry-H2B were located by correlating their position in fluorescent images on finder grids with location in the electron microscope, or in the case of serial sections, simply by correlating cell position with location on slot grids. A series of low and high magnification micrographs of regions of interest was acquired.
4.8. Overlay of fluorescence and electron images
Composite images showing the fluorescent signal overlaid on electron micrographs were created using Adobe Photoshop. As the fluorescent images were comparatively low resolution, they were upsized prior to overlaying on the electron micrograph and were then rotated and linearly scaled to fit. Each of the images was adjusted for contrast, brightness, exposure, and RGB curves/levels to more closely match the original signal as viewed on the epifluorescence microscope. Electron micrographs were adjusted linearly for brightness and contrast, and were sharpened using unsharp mask.
4.9. Spectral profiling
Spectral confocal imaging was undertaken using a Zeiss LSM 780. Samples were presented on a glass slide and imaged using a Plan Apochromat 20×/0.8 objective. For GFP samples, the 488 nm laser line was principally used for excitation via a 488 DCLP and spectral information collected every 8.7 nm from 465–692 nm. Excitation was also tested using the 458 nm laser line via the 458 DCLP filter, which showed no change in emission profile. For mCherry the 561 nm laser line was used for excitation via a 458/561 DCBP and emission collected every 8.7 nm from 465–692 nm. Importantly, for cross comparison of pre-sectioned samples to fully processed samples, the detector voltage was kept constant. GFP photobleaching was carried out by imaging over 83 sequential scans with a pixel dwell time of 1.72 μsec using 60% transmission of the 488 nm laser and emission collection between 493–560 nm. mCherry photobleaching was carried out by imaging over 83 sequential scans with a pixel dwell time of 2.08 μsec using 40% transmission of the 561 nm laser and emission collection between 578–696 nm.
4.10. Integrated light and scanning electron microscopy
The integrated light and scanning electron microscope consisted of a Quanta 250 FEG (FEI Company, Eindhoven) and attached SECOM platform with Nikon Plan Apo 40×/0.95 objective (DELMIC B.V., Delft). GFP fluorescence was stimulated by excitation of GFP using a 488 nm laser light source and multi-band filters (FF505/606-Di01 dichroic, FF01-524/628-25 emission; Semrock, Rochester NY). Images were collected using an iXon Ultra camera (Andor Technology, Belfast) with a laser power of 10 mW, exposure time of 2 s, at chamber pressures of both 200 pA and 40 pA. For comparisons across vacuum conditions, GFP fluorescence was stimulated by excitation using an LED light engine (475/28 nm; Lumencor Inc., Beaverton OR). Images were collected using a Clara CCD camera (Andor Technology, Belfast) with exposure time set at 3.9 s. When directly comparing fluorescence intensity across a range of vacuum pressures, the exposure time was kept constant, and adjustments in brightness, contrast and RGB levels were made post-acquisition to closely match the levels of background fluorescence between images. For SEM imaging, the vCD backscatter detector (FEI Company, Eindhoven) was used at a working distance of 5.9 mm, and inverted contrast images were acquired at both 40 pA, and in high vacuum (2.5 keV, spot size 3.5, 30 μm aperture, and dwell time of 60 μs for all chamber pressures). The electron micrographs were adjusted to enhance contrast, and composite images of dual signals were generated as outlined above.
Acknowledgements
The authors would like to acknowledge funding from Cancer Research UK, and from the MRC, BBSRC and EPSRC under grant award MR/K01580X/1 to POT and LC. We would like to thank Jemima Burden for useful discussions, Ian Morrison for assistance with spectral analysis and Helmut Gnaegi (Diatome Inc.) for loan of a diamond knife.
References
- 1.Polishchuk R.S., Polishchuk E.V., Marra P., Alberti S., Buccione R., Luini A., Mironov A.A. Correlative light-electron microscopy reveals the tubular-saccular ultrastructure of carriers operating between golgi apparatus and plasma membrane. J. Cell Biol. 2000;148:45–58. doi: 10.1083/jcb.148.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown E., Mantell J., Carter D., Tilly G., Verkade P. Studying intracellular transport using high-pressure freezing and correlative light electron microscopy. Semin. Cell Dev. Biol. 2009;20:910–919. doi: 10.1016/j.semcdb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Agronskaia A.V., Valentijn J.A., van Driel L.F., Schneijdenberg C.T., Humbel B.M., van Bergen en Henegouwen P.M., Verkleij A.J., Koster A.J., Gerritsen H.C. Integrated fluorescence and transmission electron microscopy. J. Struct. Biol. 2008;164:183–189. doi: 10.1016/j.jsb.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Faas F.G., Barcena M., Agronskaia A.V., Gerritsen H.C., Moscicka K.B., Diebolder C.A., van Driel L.F., Limpens R.W., Bos E., Ravelli R.B., Koning R.I., Koster A.J. Localization of fluorescently labeled structures in frozen-hydrated samples using integrated light electron microscopy. J. Struct. Biol. 2013;181:283–290. doi: 10.1016/j.jsb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Karreman M.A., Buurmans I.L., Agronskaia A.V., Geus J.W., Gerritsen H.C., Weckhuysen B.M. Probing the different life stages of a fluid catalytic cracking particle with integrated laser and electron microscopy. Chemistry. 2013;19:3846–3859. doi: 10.1002/chem.201203491. [DOI] [PubMed] [Google Scholar]
- 6.Liv N., Zonnevylle A.C., Narvaez A.C., Effting A.P., Voorneveld P.W., Lucas M.S., Hardwick J.C., Wepf R.A., Kruit P., Hoogenboom J.P. Simultaneous correlative scanning electron and high-NA fluorescence microscopy. PloS One. 2013;8:e55707. doi: 10.1371/journal.pone.0055707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zonnevylle A.C., Van Tol R.F., Liv N., Narvaez A.C., Effting A.P., Kruit P., Hoogenboom J.P. Integration of a high-NA light microscope in a scanning electron microscope. J. Microsc. 2013;252:58–70. doi: 10.1111/jmi.12071. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z., Hu B., Zhang Y., Luo Q., Gong H. Development of a plastic embedding method for large-volume and fluorescent-protein-expressing tissues. PloS One. 2013;8:e60877. doi: 10.1371/journal.pone.0060877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keene D.R., Tufa S.F., Lunstrum G.P., Holden P., Horton W.A. Confocal/TEM overlay microscopy: a simple method for correlating confocal and electron microscopy of cells expressing GFP/YFP fusion proteins. Microsc. Microanal. Off. J. Microsc. Soc. Am. Microbeam Anal. Soc. Microsc. Soc. Can. 2008;14:342–348. doi: 10.1017/S1431927608080306. [DOI] [PubMed] [Google Scholar]
- 10.Karreman M.A., Agronskaia A.V., Verkleij A.J., Cremers F.F., Gerritsen H.C., Humbel B.M. Discovery of a new RNA-containing nuclear structure in UVC-induced apoptotic cells by integrated laser electron microscopy. Biol. Cell. 2009;101:287–299. doi: 10.1042/BC20080076. [DOI] [PubMed] [Google Scholar]
- 11.Cortese K., Vicidomini G., Gagliani M.C., Boccacci P., Diaspro A., Tacchetti C. 3D HDO-CLEM: cellular compartment analysis by correlative light-electron microscopy on cryosection. Methods Cell Biol. 2012;111:95–115. doi: 10.1016/B978-0-12-416026-2.00006-6. [DOI] [PubMed] [Google Scholar]
- 12.Fabig G., Kretschmar S., Weiche S., Eberle D., Ader M., Kurth T. Labeling of ultrathin resin sections for correlative light and electron microscopy. Methods Cell Biol. 2012;111:75–93. doi: 10.1016/B978-0-12-416026-2.00005-4. [DOI] [PubMed] [Google Scholar]
- 13.Karreman M.A., Agronskaia A.V., van Donselaar E.G., Vocking K., Fereidouni F., Humbel B.M., Verrips C.T., Verkleij A.J., Gerritsen H.C. Optimizing immuno-labeling for correlative fluorescence and electron microscopy on a single specimen. J. Struct. Biol. 2012;180:382–386. doi: 10.1016/j.jsb.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Kukulski W., Schorb M., Welsch S., Picco A., Kaksonen M., Briggs J.A. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J. Cell Biol. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kukulski W., Schorb M., Kaksonen M., Briggs J.A. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 2012;150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- 16.Sims P.A., Hardin J.D. Fluorescence-integrated transmission electron microscopy images: integrating fluorescence microscopy with transmission electron microscopy. Methods Mol. Biol. (Clifton, N.J.) 2007;369:291–308. doi: 10.1007/978-1-59745-294-6_14. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe S., Punge A., Hollopeter G., Willig K.I., Hobson R.J., Davis M.W., Hell S.W., Jorgensen E.M. Protein localization in electron micrographs using fluorescence nanoscopy. Nat. Methods. 2011;8:80–84. doi: 10.1038/nmeth.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luby-Phelps K., Ning G., Fogerty J., Besharse J.C. Visualization of identified GFP-expressing cells by light and electron microscopy. J. Histochem. Cytochem. 2003;51:271–274. doi: 10.1177/002215540305100301. [DOI] [PubMed] [Google Scholar]
- 19.Nixon S.J., Webb R.I., Floetenmeyer M., Schieber N., Lo H.P., Parton R.G. A single method for cryofixation and correlative light, electron microscopy and tomography of zebrafish embryos. Traffic (Copenhagen, Denmark) 2009;10:131–136. doi: 10.1111/j.1600-0854.2008.00859.x. [DOI] [PubMed] [Google Scholar]
- 20.Fabrowski P., Necakov A.S., Mumbauer S., Loeser E., Reversi A., Streichan S., Briggs J.A., De Renzis S. Tubular endocytosis drives remodelling of the apical surface during epithelial morphogenesis in Drosophila. Nat. Commun. 2013;4:2244. doi: 10.1038/ncomms3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell K., Mitchell S., Paultre D., Posch M., Oparka K. Correlative imaging of fluorescent proteins in resin-embedded plant material. Plant Physiol. 2013;161:1595–1603. doi: 10.1104/pp.112.212365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larijani B., Barona T.M., Poccia D.L. Role for phosphatidylinositol in nuclear envelope formation. Biochem. J. 2001;356:495–501. doi: 10.1042/0264-6021:3560495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumas F., Byrne R.D., Vincent B., Hobday T.M., Poccia D.L., Larijani B. Spatial regulation of membrane fusion controlled by modification of phosphoinositides. PloS One. 2010;5:e12208. doi: 10.1371/journal.pone.0012208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domart M.C., Hobday T.M., Peddie C.J., Chung G.H., Wang A., Yeh K., Jethwa N., Zhang Q., Wakelam M.J., Woscholski R., Byrne R.D., Collinson L.M., Poccia D.L., Larijani B. Acute manipulation of diacylglycerol reveals roles in nuclear envelope assembly & endoplasmic reticulum morphology. PloS One. 2012;7:e51150. doi: 10.1371/journal.pone.0051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.P. Varnai, T. Balla, Live cell imaging of phosphoinositide dynamics with fluorescent protein domains, Biochim. Biophys. Acta, 1761 (2006) 957-967. [DOI] [PubMed]
- 26.Balla T., Varnai P. Visualizing cellular phosphoinositide pools with GFP-fused protein-modules. Curr. Protoc. Cell Biol. 2009 doi: 10.1126/stke.2002.125.pl3. (Unit 24.4) [DOI] [PubMed] [Google Scholar]
- 27.Downes C.P., Gray A., Lucocq J.M. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005;15:259–268. doi: 10.1016/j.tcb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Nickels J.D., Neill H.O., Hong L., Tyagi M., Ehlers G., Weiss K.L., Zhang Q., Yi Z., Mamontov E., Smith J.C., Sokolov A.P. Dynamics of protein and its hydration water: neutron scattering studies on fully deuterated GFP. Biophys. J. 2012;103:1566–1575. doi: 10.1016/j.bpj.2012.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald K.L., Webb R.I. Freeze substitution in 3 h or less. J. Microsc. 2011;243:227–233. doi: 10.1111/j.1365-2818.2011.03526.x. [DOI] [PubMed] [Google Scholar]
- 30.Song L., Varma C.A., Verhoeven J.W., Tanke H.J. Influence of the triplet excited state on the photobleaching kinetics of fluorescein in microscopy. Biophys. J. 1996;70:2959–2968. doi: 10.1016/S0006-3495(96)79866-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keune W., Bultsma Y., Sommer L., Jones D., Divecha N. Phosphoinositide signalling in the nucleus. Adv. Enzyme Regul. 2011;51:91–99. doi: 10.1016/j.advenzreg.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Malhas A.N., Vaux D.J. The nuclear envelope and its involvement in cellular stress responses. Biochem. Soc. Trans. 2011;39:1795–1798. doi: 10.1042/BST20110719. [DOI] [PubMed] [Google Scholar]
- 33.Wacker I., Schroeder R.R. Array tomography. J. Microsc. 2013;252:93–99. doi: 10.1111/jmi.12087. [DOI] [PubMed] [Google Scholar]
- 34.Armer H.E., Mariggi G., Png K.M., Genoud C., Monteith A.G., Bushby A.J., Gerhardt H., Collinson L.M. Imaging transient blood vessel fusion events in zebrafish by correlative volume electron microscopy. PloS One. 2009;4:e7716. doi: 10.1371/journal.pone.0007716. [DOI] [PMC free article] [PubMed] [Google Scholar]