Abstract
Pik-h, an allele of Pik, confers resistance against the rice blast pathogen Magnaporthe oryzae. Its positional cloning has shown that it comprises a pair of NBS-LRR genes, Pikh-1 and Pikh-2. While Pikh-1 appears to be constitutively transcribed, the transcript abundance of Pikh-2 responds to pathogen challenge. The Pikh-1 CC (coiled coil) domain interacts directly with both AvrPik-h and Pikh-2. Transient expression assays demonstrated that Pikh-2 mediates the initiation of the host defence response. Nucleocytoplasmic partitioning of both Pikh-1 and Pikh-2 is required for their functionalities. In a proposed mechanistic model of Pik-h resistance, it is suggested that Pikh-1 acts as an adaptor between AvrPik-h and Pikh-2, while Pikh-2 transduces the signal to trigger Pik-h-specific resistance.
Introduction
The surveillance system evolved by plants to detect and defend against microbial attack requires a capacity to recognize a plethora of pathogens. The result of detection and successful defense is often the hypersensitive response (HR; [1]), which is triggered when a pathogen's avirulence (Avr) gene is recognized by a matching host resistance (R) gene. The genetic relationship between the R and Avr genes was elegantly interpreted by Flor in 1971, and modeled in his “gene-for-gene” hypothesis [2]. However, genome-wide analyses have increasingly revealed that the host R gene repertoires are far smaller than what would be needed to respond specifically to every pathogen that it may encounter [3], [4]. This fact implies that individual R genes must have the capacity to recognize more than one pathogen race and even family [5], [6]. Although some broad-spectrum recognition mechanisms not involving R genes have been described, multiple recognition specificity is considered necessary for the perception by R genes of a wide variety of pathogens [6]–[8]. The co-evolution of host and pathogen drives the mutation of Avr proteins to alter their recognition properties, with allelic diversity at R genes representing the response [9], [10].
Numerous R genes have been isolated in recent years. Most have proven to encode a large cytoplasmic protein comprising either an N-terminal coiled coil (CC) or a Toll-interleukin receptor (TIR) domain, a central nucleotide-binding site (NBS) domain, and a C-terminal leucine-rich repeat region (LRR) [11]. The NBS domain shares a degree of sequence similarity with mammalian apoptosis-inducing effectors, and its function has therefore been considered to be a switch regulating R protein activity. The LRR region is generally thought to represent the major determinant of specificity [12]. The TIR/CC domain was initially proposed to function in downstream signalling, as shown in flax L10 and Arabidopsis thaliana RPP1 [13], [14]. However, accumulating evidence also supported the role of this domain in effector recognition, including the cases of the A. thaliana RPS5, the tobacco N proteins [15], [16], and the potato RB proteins [8]. Therefore, the TIR/CC domain has become the subject of intense interest for its dual functions in “gene-for-gene” interactions. Recently, the importance of this domain for R protein function was further highlighted in two studies, which demonstrated that TIR/CC domain-dependent dimerization is required for downstream immune signaling [17], [18].
Plant R proteins have been found to locate in diverse sub-cellular compartments. In the past few years, a number of studies have situated the nucleus in the center of attention for disease resistance activation. For instance, upon elicitor induction, several R proteins, including the tobacco N, the barley MLA10, and the A. thaliana RPS4 and SNC1 proteins, were found to accumulate in the nucleus, and this nuclear accumulation is required for their proper functioning [19]–[23]. However, two recent studies demonstrated that the nucleocytoplasmic potato Rx protein requires both nuclear and cytoplasmic pools for its activation [24], [25]. Apart from nucleocytoplasmic distribution, R proteins may also be found in the plasma membrane, as is the case with A. thaliana RPM1 [26]. Through nuclear exclusion and direct membrane tethering, Gao et al. showed that activation and signalling of RPM1 occurs at the plasma membrane and initiates a cytosolic pathway [26]. Therefore, plant R proteins can function in specific cellular compartments depending on the protein.
Some examples have emerged where resistance against a single pathogen requires not just one, but a pair of NBS-LRR genes [27]. The earliest example is related to resistance against Peronospora parasitica in A. thaliana [28], but subsequently, the same scenario has been established for a viral resistance in tobacco [29], leaf rust resistance in wheat [30], blast resistance in rice [31]–[35], and the resistance against three distinct pathogens in A. thaliana [4]. As yet it is still unclear how such coupled genes can act co-operatively to establish effector recognition, but detailed examination of the Pik locus in rice has shown that resistance does indeed require functional versions of both genes to be present.
Here we report the function of and the nature of the interaction between the coupled genes responsible for Pik-h encoded rice blast resistance. Genetic analysis has established that a gene-for-gene interaction exists between various AvrPik and Pik alleles [34]–[37]. Here, we have elaborated a working model for the AvrPik-h and Pik-h interaction, in which one of the R gene products (Pikh-1) acts as an adaptor between AvrPik-h and the other R gene product (Pikh-2), while Pikh-2 transduces the signal to trigger the Pik-h-specific resistance.
Results
Identification and Validation of Candidates for Pik-h
The Pik-h donor cultivar (cv.) K3 was crossed with the blast susceptible cv. As20-1, and Pik-h resistance in the resulting F2 population segregated in a monogenic fashion [resistant/susceptible: 484/150 (χc 2 = 0.54; P>0.40)]. Linkage analysis placed Pik-h within a 237 kb interval flanked by markers K37 (proximal) and K28 (distal), a location which is identical to that of both Pik and Pik-p, in which two haplotypes K and N have been shown to differ by a large insertion/deletion (Table S1; Figure S1; [34], [35]).
Since cvs K3 (Pik-h carrier), K60 (Pik-p) and Kusabue (Pik) all possess the K haplotype [35], the Pikp-1/Pik-1 pair (for simplicity, the K1 gene) and the Pikp-2/Pik-2 pair (K2 gene) were considered to be the most likely candidates for Pik-h (Figure S1). A gain-of-function test based on the introduction into susceptible cvs of one of three distinct constructs- the first harboring Pikh-1 alone, the second Pikh-2 alone and the third both Pikh-1 and Pikh-2 (referred to as Pikh-12)- was performed. Transgenic plants carrying Pikh-1 or Pikh-2 constructs alone did not gain a complementary phenotype, while those carrying the Pikh-12 construct in susceptible backgrounds did (Table 1; Figure S2), indicating that the expression of Pik-h resistance requires two independent genetic determinants. Loss-of-function tests of Pikh-1 and Pikh-2 independently further confirmed that both genes were involved in Pikh-mediated blast resistance (Table 1; Figure S2).
Table 1. Characterization of the coupled genes for Pik-h via gain and loss of function approaches.
| Recipient | Inoculation | Reaction of T0 Plants a | Success | |||||
| Candidate/construct | Cultivar | Isolate | R | MR | MS | S | Total | ratio (%) b |
| Gain of function | ||||||||
| Pikh-1 | Q1063 | CHL346 | 0 | 0 | 1 | 149 | 150 | 0 |
| Pikh-2 | Q1063 | CHL346 | 0 | 0 | 0 | 82 | 82 | 0 |
| Pikh-12 | Kuyuku 131 | CHL42 | 7 | 3 | 1 | 8 | 19 | 42.1 |
| Pikh-12 | K60 | CHL42 | 82 | 14 | 14 | 4 | 114 | 84.2 |
| Loss of function c | ||||||||
| Pikh-1 RNAi (KP3i1) | K3 | CHL346 | 21 | 0 | 4 | 17 | 42 | 50.0 |
| Pikh-2 RNAi (KP4i) | K3 | CHL346 | 5 | 0 | 0 | 20 | 25 | 80.0 |
T0 plants derived from each construct were inoculated with the Pikh-avirulent isolates, CHL346 or CHL42. R, resistant; MR, moderate resistant; MS, moderate susceptible; S, susceptible.
The success ratios for gain- and loss-of-function complementation tests were calculated as (R+MR)/(R+MR+MS+S), and (MS+S)/(R+MR+MS+S), respectively.
The Pik allele-specific RNAi constructs, KP3 RNAi1 and KP4 RNAi, which correspond to Pikp-1 and Pikp-2, respectively [40], were transformed into the Pik-h carrier cv K3.
Sequence data from this article can be found in the GenBank/EMBL databases under accession numbers HQ662330 (Pik-h).
Transcription Profiles of Pikh-1 and Pikh-2
The transcript abundance of Pikh-1 in plants infected by an avirulent race of rice blast fell over the first 12 h following inoculation, but then increased steadily to its constitutive level by 72 h. Since the same profile was observed in the mock-inoculated control (Figure 1), this response was most likely associated with the inoculation procedure itself rather than to an interaction with the pathogen. However, the transcription of Pikh-2 rose after 12 h, reaching about two fold of the constitutive level by 72 h after inoculation. Given the different expression pattern of Pikh-1 and Pikh-2, it is tempting to speculate that the coupled genes were not co-regulated upon the pathogen challenge.
Figure 1. Transcription profiles of the coupled genes Pikh-1 and Pikh-2, as assayed by qRT-PCR.
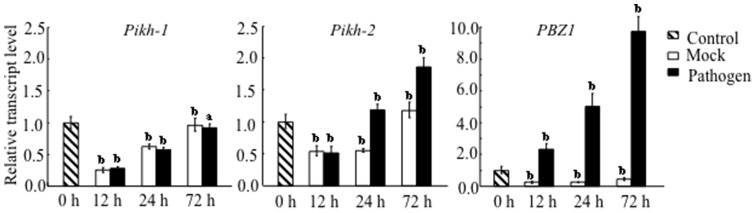
Data represent mean ± standard deviation (n = 3). Similar results were obtained from two independent experiments. Lower case letter (a, b) used to indicate whether the treatment and control means differed at P<0.01 or P<0.05 level, respectively. The pathogen-inducible gene PBZ1 [46] was used as a reference.
Interactions among Pikh-1, Pikh-2 and AvrPik-h in Vitro
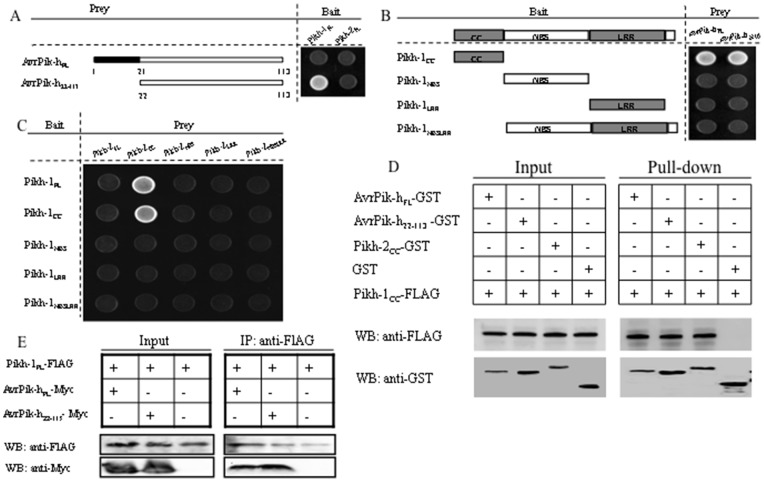
Yeast two-hybrid assays demonstrated that the full-length Pikh-1 (Pikh-1FL) protein, but not Pikh-2, interacted with the cognate mature protein, AvrPik-h22–113 (a signal peptide truncated form of AvrPik-h, identical to AvrPik-D [37]) (Figure 2A). The implication was that Pikh-2 is unlikely to be receptor for effector recognition. When a series of truncated Pikh-1 proteins was tested for interaction with both the full-length and the truncated forms of AvrPik-h, it was apparent that the CC domain (Pikh-1CC) was both necessary and sufficient for the interaction to occur (Figure 2B). Interactions were also detected between Pikh-2CC and both Pikh-1FL and Pikh-1CC (Figure 2C). These interactions were verified by the pull-down and co-immnoprecipitation assays (Figures 2D and 2E). The expression of all bait and prey proteins was confirmed by Western blot analysis (Figure S4A).
Figure 2. Interactions among Pikh-1, Pikh-2 and AvrPik-h proteins as detected by the yeast two hybrid, pull-down, and co-immunoprecipitation systems.
(A) Pikh-1FL and Pikh-2FL vs full length or truncated AvrPik-h proteins. (B) Defining the Pikh-1 domain interacting with AvrPik-hFL and AvrPik-h22–113. (C) Full length and truncated Pikh-1 vs Pikh-2. (D) Pikh-1CC vs AvrPik-hFL, AvrPik-h22–113 and Pikh-2CC using the pull-down assay. (E) Pikh-1FL vs AvrPik-hFLand AvrPik-h22–113 using the co-immunoprecipitation system. Western blots employed anti-FLAG (Pikh-1CC and Pikh-1FL), anti-GST (AvrPik-hFL, AvrPik-h22–113 and Pikh-2CC), and anti-Myc (AvrPik-hFL and AvrPik-h22–113) antibodies.
Interactions among Pikh-1, Pikh-2 and AvrPik-h in vivo
When GFP-fused proteins consisting of either the full-length or isolated domains of the three genes were separately expressed in rice protoplasts, distinct distributions of GFP signal were observed. The Pikh-1FL-GFP construct demonstrated a balanced distribution between cytoplasm and nucleus, whereas the Pikh-1CC-GFP exclusively accumulated in nucleus. A higher level of fluorescence signal of the Pikh-1NBS-GFP was detected in the nucleus than in cytoplasm, while the Pikh-1LRR-GFP was excluded from the nucleus and distributed throughout the cytoplasm (Figure S5A). In contrast, both Pikh-2FL-GFP and Pikh-2CC-GFP accumulated in both the nucleus and the cytoplasm, and both Pikh-2NBS-GFP and Pikh-2LRR-GFP exclusively, in the nucleus (Figure S5B). Additionally, both AvrPik-hFL-GFP and AvrPik-h22–113-GFP distributed between cytoplasm and nucleus (Figure S5C). As expected, the nuclear localization signal (NLS) fused versions of both Pikh-1FL-GFP and Pikh-2FL-GFP accumulated in the nucleus, while the nuclear export signal (NES) carrying versions of both the full-length and CC domain of the coupled genes excluded from the nucleus (Figure S5D).
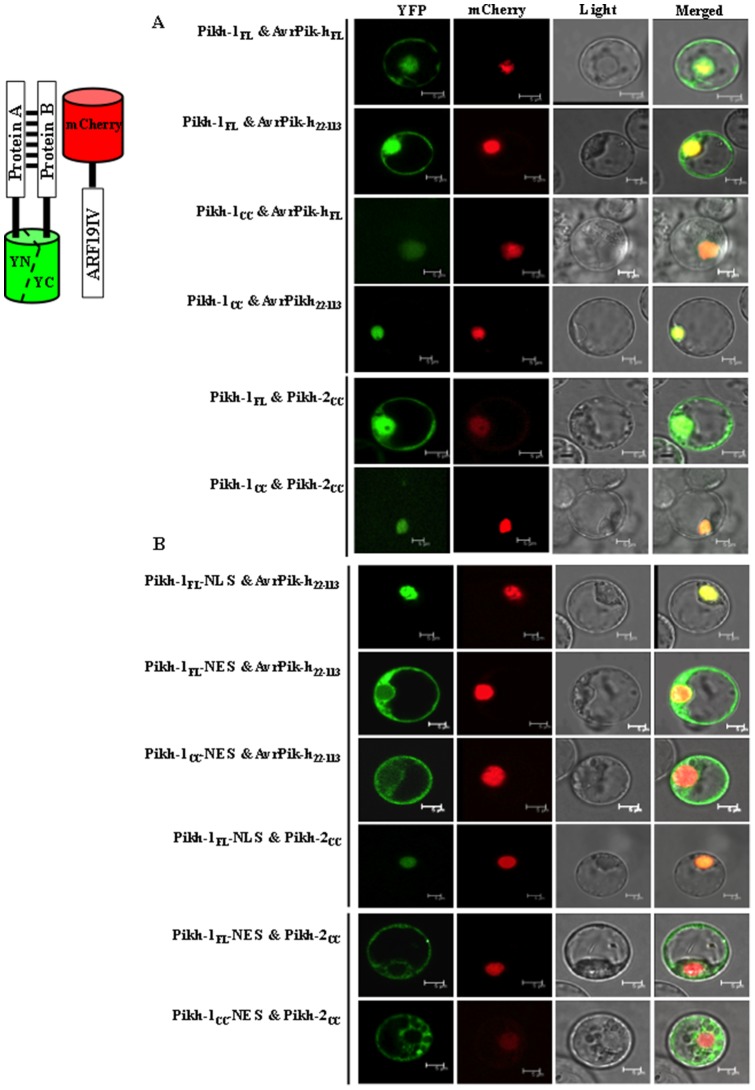
The positive interactions observed in vitro were further confirmed by the bimolecular fluorescence complementation (BiFC) assay in a rice protoplast system. The localization and distribution of all the combinations of the triple proteins was stable, but not dynamic between cellular organs. That is, The Pikh-1FL protein formed a complex with either AvrPik-hFL or AvrPik-h22–113 in both the cytoplasm and the nucleus, but the Pikh-1CC altered this nucleocytoplasmic partitioning to nucleus, only (Figure 3A). The Pikh-1FL protein also formed a complex with Pikh-2cc in both the cytoplasm and nucleus, and once again its replacement with Pikh-1CC also disrupted the nucleocytoplasmic distribution of the Pikh-1 interaction with Pikh-2 (Figure 3A). When the NES versions of both Pikh-1FL and Pikh-1CC were co-expressed with the AvrPik-h22–113 or Pikh-2 CC, fluorescence was detected only in the cytoplasm, while the NLS versions were detected exclusively in the nucleus (Figure 3). As the negative controls, neither Pikh-1FL nor AvrPik-hFL gave BiFC with Pikh-2FL (Figure S6, upper panel). In addition, co-expression of various versions of Pikh-1, Pikh-2, and AvrPik-h proteins with the non-allelic blast R gene protein, Pib, did not produce any fluorescence (Figure S4B; and Figure S6, lower panel). Moreover, a nucleocytoplasmic fractionation-based Western blot assay was applied to assure the localizations of interactions of Pikh-1FL and Pikh-1CC with AvrPik-h22–113. As expected, the former combination was detected in both nuclear and the nuclei-depleted fractions, and the latter one was detected only in the nuclei fraction (Figure S7). Together, these indicated that the fluorescence detected was due to a specific association of the proteins related.
Figure 3. Interactions among Pikh-1, Pikh-2, and AvrPik-h proteins as detected by bimolecular fluorescence complementation (BiFC) assay.
The images shown are representative of at least three independent experiments. (A) Pikh-1FL complexed with AvrPik-hFL, AvrPik-h22–113, and Pikh-2CC in both the nucleus and the cytoplasm, while Pikh-1CC produces such interactions only in the nucleus. (B) The targeting signals NLS and NES redirected interactions to, respectively, the nucleus and the cytoplasm. ARF19IV-mCherry, nuclear marker.
Functionalities upon interactions among Pikh-1, Pikh-2 and AvrPik-h
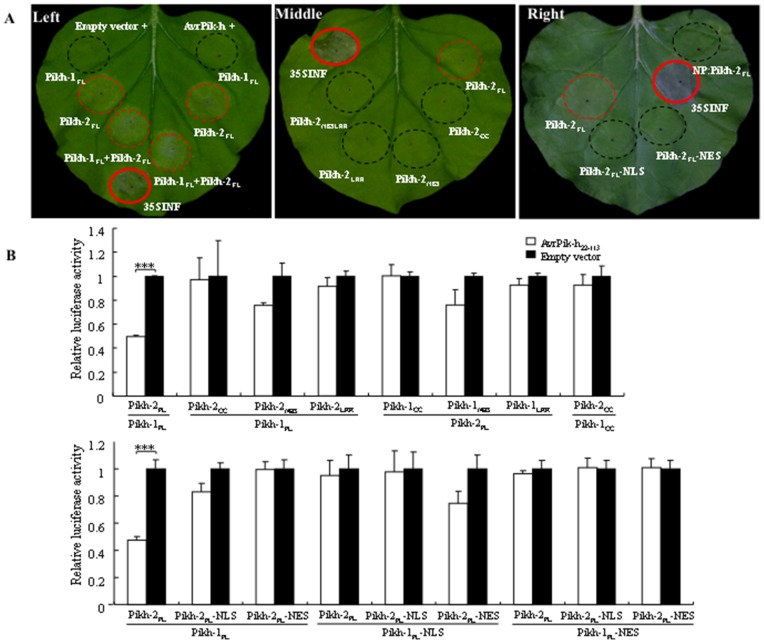
The molecular basis for Pik-h activation was investigated via an Agrobacterium-mediated transient expression system in Nicotiana benthamiana, in which an HR-like response was induced by the co-expression of various combinations of the Pik-h and AvrPik-h proteins. All combinations involving Pikh-2FL induced an HR-like response, demonstrating that it could be auto-activated under the control of the CaMV 35S promoter (Figure 4A, left panel). The analysis of truncated Pikh-2 derivatives showed that only the Pikh-2FL protein was able to induce HR (Figure 4A, middle panel). Since the Pikh-2FL is deposited in both the nucleus and cytoplasm (Figure S5B), a pair of constructs of Pikh-2FL fused to either NLS or NES was generated to test whether the subcellular distribution of Pikh-2 is of any importance for its autoactivity. This experiment showed that the restriction of Pikh-2 within either the nucleus or cytoplasm blocked its HR-triggering activity (Figure 4A, right panel; Figure S5D), which implied that both nuclear and cytoplasmic pools of Pikh-2 are needed for normal function. The expression of the Pikh-2FL protein driven by its native promoter (NP) likewise induced no HR (Figure 4A, right panel). Immunoblot analysis revealed that except for the Pikh-2 protein driven by NP, the proteins all accumulated at a comparable level (Figure S4C). Thus, the HR induction shown by the Pikh-2 protein driven by the CaMV 35S promoter may have derived from its higher level of accumulation.
Figure 4. Functionality upon interactions among Pikh-1, Pikh-2, and AvrPik-h proteins.
(A) Transient expression in N. benthamiana. Pikh-2 on its own triggered an HR-like response (left). The Pik-h domains required for the HR (middle). The response induced by various versions of Pikh-2 (right). The NES or NLS redirected distributions of Pikh-2 resulted in the loss of its functionality. NP: Native promoter. The p35S-INF construct was used as positive control. (B) Transient assay of HR in cv. Nipponbare protoplasts using firefly luciferase as a reporter. The R gene constructs were driven by their NP, and Avr by the maize ubiquitin promoter. The reduction in luminescence was compared with the control, where an empty vector, instead of AvrPik-h22–113, was uesd for protoplast transformation. Each assay consisted of three technical and three biological replicates. ***P<0.001. Co-expression of the full length Pikh-1 and Pikh-2 was necessary to confer AvrPik-h-dependent HR, and the NES or NLS redirected subcellular distributions of Pikh-1 and Pikh-2 resulted in loss of functionality.
Since auto-activation has been observed in heterologous plant expression systems, a firefly luciferase reporter gene-based HR-like cell death assay was performed in rice protoplasts to investigate how Pik-h might function. As the full-length AvrPik-h protein displayed auto-activation, while the signal peptide truncated construct ubi::AvrPik-h22–113 did not (Figure S8), the latter was challenged by both Pikh-1 and Pikh-2 expressed by their NP (Figure 4B, upper panel). Only the combination of AvrPik-h22–113 and Pikh-1FL and Pikh-2FL resulted in a significant reduction in luciferase activity. In contrast, the combination among AvrPik-h22–113 and Pikh-1FL protein with any Pikh-2 truncated protein, or AvrPik-h22–113 and Pikh-1 truncated protein with the Pikh-2FL protein did not induce significant HR-like cell death in rice protoplasts. Interestingly, combination of AvrPik-h22–113, and the CC domains of both Pikh-1 and Pikh-2 did not cause significant cell death, indicating that the parsimonious regions involved in the protein-protein interactions aforementioned were insufficient to trigger the effector-dependent HR.
To further determine whether the nucleocytoplasmic partitioning was required for the Pikh-mediated resistance, both Pikh-1 and Pikh-2 were fused either to NLS or NES and expressed under the control of their NP in rice protoplasts. Combinations involving AvrPik-h22–113 and the NLS or NES versions of Pikh-1FL with any of the Pikh-2 versions induced no HR-like cell death and vice versa (Figure 4B, lower panel), even though the NLS- or NES-tagged proteins were as abundant as the wild types (Figure S4D). Hence, it is tempting to speculate that the balanced distribution of Pikh-1 and Pikh-2 in both nucleus and cytoplasm is essential for the Pikh-mediated resistance.
Discussion
Positional divergence of the coupled genes for the Pik alleles
Although the majority of R proteins share a tripartite structure consisting of a CC/TIR, an NBS domain and an LRR region, the role of each of these components is no longer as clear cut as was once believed. The NBS domain was initially thought to regulate the activation of the R protein [38], but the observation that the expression of just the NBS domain of the potato Rx protein on its own is sufficient to trigger a constitutive defence response has led to the modified theory that it can also serve as an interaction platform for studying downstream signalling components [11], [39]. The LRR region was generally held to be responsible for Avr perception, as it has clearly been subjected to strong diversifying selection. Compelling evidence for this role has been provided by both in vitro or in vivo assays of the rice Pita and the A. thaliana RPP1 proteins [14], [40], where in both cases the LRR region was necessary and sufficient for the R/Avr interaction. The supposed function of the CC/TIR domain was associated with downstream signalling [13], [14], but it has become clear that it is also involved with pathogen recognition [15], [16]. According to the previous studies, all of the CC/TIR domains which mediate the R/Avr interaction were assembled in those R proteins being interacted with Avr proteins, indirectly, compared with the LRR domain in those R proteins interacting with Avr proteins, directly [15], [15,41]. However, it is the CC domain of Pikh-1 which is necessary and sufficient for direct binding with AvrPik-h (Figures 2B and 2D), consistent with the observation that the variable amino acids in Pikh-1 were found mostly within its CC domain (Figure S3A). The present results supply a clear example for the mediation of pathogen perception by direct interaction with an N-terminal domain. The Pikh-1 CC domain was also responsible for the interaction between Pikh-1 and Pikh-2 (Figures 2C and 2D), implying that it may act as a molecular bridge relaying a signal from AvrPik-h to Pikh-2. It has been reported that self-association of the L6 TIR domain, which could provide a scaffold for downstream signaling protein binding, is a requirement for immune activation [17]. Similarly, the crystal structure of the MLA CC domain has revealed that the CC dimer serves as a minimal functional module in cell death initiation [18]. The CC domain-dependent homodimerization of MLA has been suggested to attract particular WRKY hetero- or homo-oligomers for the purpose of downstream signaling [18]. Whether the heterodimer of the CC/TIR domain is therefore also a critical player in downstream signaling remains an open question. However, the co-expression of the CC domains of both Pikh-1 and Pikh-2 was insufficient to induce HR (Figure 4B), suggesting that the association of the two CC domains may be related to the formation of AvrPikh-Pikh1-Pikh2 recognition complexes rather than specifically related to a downstream signaling event.
The sub-cellular localization of R proteins is relevant to their activity in resistance signalling. The tobacco R gene product N, which confers resistance against tobacco mosaic virus (TMV), is found in both the cytoplasm and the nucleus. During TMV infection, the shuttling of p50-activated N from the cytoplasm to the nucleus appears to be required for an effective defence response [19]. As a second example, the activity of the A. thaliana SNC1 and RPS4 have also been associated with their nuclear accumulation [21], [22]. Strikingly, a recent study has confirmed that the enhanced defense responses mounted under conditions of ABA deficiency and high temperature is dependent on the nuclear localization of SNC1 and RPS4 [23]. In contrast, the regulation of the potato CC-NBS-LRR Rx protein (which confers resistance against potato virus X) requires its nucleocytoplasmic distribution [24], [25], which is brought about by its CC and LRR domains and facilitated by an accessory protein. While the nuclear Rx protein may play a role in transcriptional reprogramming, the cytoplasmic form mounts a defence process against the virus [24], [25]. With respect to the Pik-h product, the Pikh-1 CC domain disrupted its balanced nucleocytoplasmic partitioning through its interaction with both AvrPik-h and Pikh-2 (Figure 3A). When GFP fusion proteins comprising either the full-length Pikh-1 or specific Pikh-1 domains were expressed in rice protoplasts, it was only the former which retained a balanced distribution between the cytoplasm and the nucleus (Figure S5A). Thus, similar to the situation with Rx, the Pikh-1 CC and LRR domains may well have contrasting roles in determining the cellular distribution of the full-length Pikh-1 protein. Given that the full-length Pikh-1 protein was required for HR induction (Figure 4B), and that it interacted with AvrPik-h or Pikh-2 in both nucleus and cytoplasm (Figure 3A), the assumption is that Pikh-mediated resistance relies on its balanced cellular distribution. Indeed, the enforced restriction of Pikh-1 to either the nucleus or the cytoplasm blocked the AvrPikh-dependent cell death (Figure 4B). The full-length and CC domain of Pikh-2 accumulated mostly, and the NBS and LRR domains exclusively, in the nucleus (Figure S5B). An enhanced nuclear or cytoplasmic accumulation of Pikh-2 also compromised its HR activity (Figures 4A and 4B). The inference is that in the expression of Pik-h resistance, Pikh-2 may interact with Pikh-1 in the nucleus to direct transcriptional reprogramming, and in the cytoplasm to recruit certain critical downstream signaling machineries.
Functional divergence of the coupled Pik genes
Characterizing the function of each constituent of a coupled gene system like Pik-h would represent a major step towards understanding the mechanisms of R gene systems. The transcription profiles of Pikh-1 and Pikh-2 clearly differ from one another, with the former being constitutively expressed and the latter responding to pathogen challenge (Figure 1). A possible scenario is that the constitutive expression of Pikh-1 is required for elicitor recognition, whereas the inducible expression of Pikh-2 reflects its participation in downstream signalling following pathogen infection. The coupled genes (Pi5-1 and Pi5-2) responsible for Pi5-encoded blast resistance have similar transcription profiles with that of Pikh-1 and Pikh-2, respectively [32], which may represent a mechanism shared by a number of the dual R-genes systems. Pik-1 sequences were more divergent than those of Pik-2 (Figures S3A and S3B), which suggests that positive selection has been operating more heavily on the former alleles. The induction of HR-like cell death in N. benthamiana required the presence of the full length Pikh-2 protein (Figure 4A), and this was also needed to induce HR-like cell death in rice protoplasts in the presence of both Pikh-1 and AvrPik-h (Figure 4B). The over-expression of NRG1, a signalling component of the defence response, causes ectopic cell death in the absence of its partner, N [29]. In line with this observation, Pikh-2 (rather than Pikh-1) appears to be the protein responsible for the activation of downstream signalling. Finally, Pikh-2 was found to interact directly with Pikh-1, but not with AvrPik-h (Figure 2).
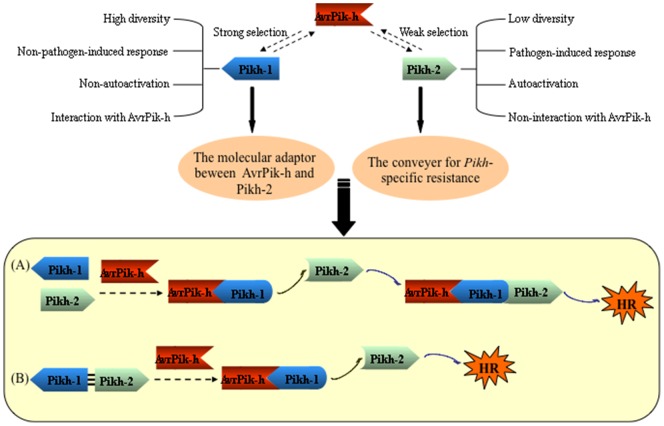
Two leading models have been elaborated to explain how a single plant R protein responds to its cognate pathogen effector. In the ligand-receptor model, recognition occurs as a result of a direct physical association between the Avr and the R proteins [9], [40], [42], [43]. In the second model, the R protein recognizes the matching Avr in a more mechanistically complex fashion, involving an accessory protein [44]. The Avr effector modifies the accessory protein in a way which the R protein perceives as a signal, and this then triggers the defence response. The association of the R protein with the accessory protein can be either constitutive [15], or can occur only after the Avr-induced modification of the accessory protein [16]. When two R genes are required for resistance, the likely situation is that the more parsimonious strategy is adopted, in which one gene is involved in elicitor recognition via direct interaction, and the other acts as a downstream component. In the case of the Pik-h resistance, Pikh-1 appears to represent a molecular adaptor between AvrPik-h and Pikh-2, while Pikh-2 is a genuine conveyer for the Pik-h-specific resistance. Two possible mechanisms underlying Pik-h activation are illustrated in Figure 5. In the absence of the Avr effector, both Pikh-1 and Pikh-2 are maintained in an inactive state. Once the presence of AvrPik-h has been detected, Pikh-1 adopts a signalling-competent conformation, which relays a signal to Pikh-2, inducing a conformational change. The consequent activation of Pikh-2 allows it to recruit specific transcription factors in the nucleus and the necessary downstream signaling proteins in the cytoplasm, thereby triggering the defence response (Figure 5A). Alternatively, Pikh-1 is constitutively associated with Pikh-2 in planta. Recognition of AvrPik-h de-stabilizes this association, which releases Pikh-2 from its structural constraints. The unconstrained conformation of Pikh-2 unlocks its signalling potential, resulting in the initiation of the necessary transcriptional reprogramming and the recruitment of the critical downstream signaling machinery (Figure 5B; [28]). The identification of the signal transduction pathway and the definition of its functional structures have become the focus of our continuing research into this important crop plant R gene.
Figure 5. A proposed model for the function of the coupled Pik-h genes.
(A) Pikh-1 perceives the Avr signal and relays it to Pikh-2, whereupon Pikh-2 defence response. (B) Pikh-1 is constitutively associated with Pikh-2. The recognition of AvrPik-h destabilizes this association, thereby lifting the constraints on the conformation of Pikh-2, and restoring its signalling potential.
While this paper was under preparation, Kanzaki et al. [37], who reported that the binding specificity between Pik-1 and AvrPik allelic proteins correlates with the recognition (race) specificity between Pik and AvrPik alleles. We, however, cannot verify the relationships with the Pik-1 allelic proteins against the AvrPik allelic proteins. As shown in Figure S9, physical interactions were also observed between all the five Pik-1 alleles (Piks-1, Pikp-1, Pik-1, Pikm-1, and Pikh-1, considering either the full-length protein, the CC domain, and the RAXT1 motif) and three AvrPik-h alleles, AvrPik-D (AvrPik-h), AvrPik-E, and AvrPik-A [37], as well as Pikh-2CC (the Pik-2 alleles all share an identical CC domain; Figure S3B). This suggests that the Pik-1 alleles show no differential binding specificity toward either AvrPik-h or any of the Pikh-2 alleles. The non-differential binding between Pikh-1 and AvrPik-h allelic proteins may be due to the presence of the RATX1 motifs in the Pik-1 CC domains (Figure S3A), which have been recognized as an Avr recognition motif [6]. The Pik-1 protein, in short, interacts with not only AvrPik, but also AvrPia proteins [6]. Similarly, there was also no binding specificity between 17 CC domains present in the potato RB protein derived from ten wild species (amino acid sequence similarities between 65.9% and 98.2%) and the relevant P. infestans effector family IPI-O1 (avirulent) and IPI-O4 (virulent) [8]. Such results raise the question of how recognition (race) specificity relates to binding specificity.
Materials and Methods
High-resolution genetic mapping
A mapping population consists of 634 F2 progeny of the cross between the Pik-h donor cv. K3 and the blast susceptible cv. As20-1. The rough location of the R gene was initially identified using bulk segregant analysis [45], and then mapped more precisely using a set of genomic position-ready markers previously used for characterizing the Pik-p/Pik region [34], [35].
Complementation analysis
The cloning strategy applied for Pik-p and Pik [34], [35] was adopted to form constructs for the candidates Pikh-1 and Pikh-2, along with a fused candidate Pikh-12. All the constructs were validated by sequencing before being introduced as transgenes into the blast susceptible recipient cvs Q1063, Kuyuku131 and K60. To test for loss-of-function, two Pik allele-specific RNAi constructs (KP3 RNAi1 and KP4 RNAi), which target, respectively, Pikp-1 and Pikp-2 [34], were transformed into the Pik-h carrier cv. K3. Transgenic progeny were tested for reaction to blast infection with isolate CHL346 (virulent to cv. Q1063, and avirulent to cv. K3) or CHL42 (virulent to cvs Kuyuku 131 and K60, and avirulent to cv. K3), and their transgenic status was confirmed using a PCR-based assay directed against the respective construct, as described previously [35].
Transcription analysis
Seedlings of cv. K3 grown to the three leaf stage in a pathogen-free room at 25°C were inoculated with either blast isolate CHL346 or water (mock inoculation, also called pathogen infection control), and held in the dark at 25°C under 100% relative humidity for 20 h. Since the Pik alleles were sensitive to environmental conditions, such as temperature, light, and humidity, seedlings being grown in a pathogen-free tissue room were used as an environmental control (34). The Pik-h transcription profile was obtained using quantitative RT-PCR (qRT-PCR), as described previously [35]. In brief, total RNA was treated with RNase-free DNase I (Promega, Madison, WI, USA), and reverse transcribed by M-MLV (Promega). The quality of RNA was assessed by measuring absorbance at 230, 260 and 280 nm, respectively, with a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Integrity of the RNA was verified by gel electrophoresis on an ethidium bromide-stained 1% agarose-TBE gel and denaturing agarose-MOPS gel. Only the RNA samples with A260/A280 ratios between 1.8 and 2.1 and A260/A230 ratios higher than 2.0, as well as the both 28S and 18S ribosomal RNA bands with a density ratio about 2∶1 were subjected to qRT-PCR analysis. qRT-PCR was performed using SYBR Premix Ex Taq (TaKaRa, Dalian, China), on a Bio-Rad CFX96 Real-Time PCR Detection System device (Bio-Rad, Hercules, CA, USA). The rice housekeeping gene encoding Actin, which was used for the expression analyses of Pik-m (31) and Pik-p (34), was used as an endogenous control to normalize the RNA expression in each qRT-PCR. The Pik allele-specific primer sets RRT5 and RRT17 (31), respectively, were employed to detect Pikh-1 and Pikh-2 expression. Primer efficiency was verified using standard curves generated by plotting the amount of the mixed cDNA against the threshold cycle (Ct) value for a series of dilutions (six orders of magnitude). Expression of genes was calculated via the 2−ΔΔCt method. Two biological and three technical replicates were included. The pathogen-inducible gene PBZ1 [31], [46] was used as a reference. Statistical comparisons were conducted with ANOVA test using SPSS v16.0 software (http://www-01.ibm.com/software/analytics/spss/).
Yeast two hybrid analysis
The full-length and truncated cDNAs corresponding to Pikh-1, Pikh-2 and AvrPik-h (AB498875.1; [47]) were cloned in frame with the GAL4 DNA binding domain (BD) of the bait vector pGBKT7, and the GAL4 activation domain (AD) of the prey vector pGADT7, respectively. The joint transformation of a BD and an AD construct into Y2HGold yeast cells was performed using the Matchmaker Gold yeast two hybrid system (Clontech, CA. USA), following the manufacturer's instructions. Yeast transformants were grown on minimal media lacking leucine and tryptophan to select for the presence of both constructs, and potential protein-protein interactions were confirmed by growing the cells in selective minimal media lacking leucine, tryptophan, histidine and adenine. Standard positive (pGBKT7-53 and pGADT7-T) and negative (pGBKT7-Lam and pGADT7-T) controls were included in each experiment. In addition, BD constructs of the five Pik-1 alleles were also generated and subjected to test for the specificity of interaction as described above. Yeast proteins tested were extracted by the trichloro-acetic acid method, separated by SDS-PAGE, and transferred to nitrocellulose membranes (Bio-Rad) by electroblotting in a Bio-Rad Mini Trans-Blot apparatus. Membranes were blocked with 3% w/v skimmed milk, and probed with an anti-Myc or an anti-HA monoclonal antibody (Sigma, MO, USA) followed by detection with anti-rabbit IgG HRP conjugates (Bio-Rad). Labeling was detected with the ECL plus Western blotting detection system (GE Healthcare, UK).
In vitro GST pull-down assay
The CC domain of Pikh-1 was inserted into the pF3A WG vector fused to a FLAG tag construct (Promega), while the Pikh-2CC, and intact and truncated fragments of AvrPik-h were inserted into the pGEX-6P-1 vector to form a GST-fusion construct (GE Healthcare). The GST-fused proteins expressed in E. coli strain BL21 were used to pull down the FLAG-fused proteins synthesized in a wheat germ-derived in vitro transcription and translation system (Promega) using a MagneGST pull-down system (Promega), following the manufacturer's protocol. The proteins eluted from the beads were separated through a 12% SDS-PAGE gel and transferred to a nitrocellulose membrane (Bio-Rad). Protein blots were blocked with 3% w/v skimmed milk and then probed with either an anti-GST or an anti-FLAG monoclonal antibody (Sigma). Either goat anti-mouse (Sigma) or anti-rabbit IgG HRP conjugates (Bio-Rad) was used as the secondary antibody for subsequent detection via enhanced chemiluminescence (GE Healthcare). As an additional recognition specificity assay, two sets of the five Pik alleles, three versions of AvrPik-h alleles, as well as the Pikh-2CC (the Pik-2 alleles share an identical CC domain) were tested by the above procedure.
Co-immunoprecipitation assay
For the coimmunoprecipitation and immunoblot assay, Pikh-1, and AvrPik-h and AvrPik-h22–113 fragments were ligated into the binary vectors, pCXSN-FLAG and pCXSN-Myc [48], respectively. Cultures of Agrobacterium tumefaciens strain GV3101 carrying one of these constructs were incubated at 28°C overnight in LB medium containing the appropriate antibiotic selective agent. When the culture OD600 had reached ∼1.0, the cells were pelleted and re-suspended in 10 mM MgCl2, 10 mM MES, 200 µM acetosyringone, the OD600 was adjusted to 1.5, and the cells then held at room temperature for 3 h. The cultures were combined in an equimolar mixture, and infiltrated into the leaves of three-week old N. benthamiana plants using a 1 ml needle-less syringe. Leaf samples were collected two days after infiltration, and then homogenized with lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM KCl, 2.5 mM MgCl2, 0.1% v/v NP-40, 5 mM DTT, and a protease inhibitor cocktail [Roche, IN, USA]), and centrifuged at 15,000 rpm for 30 min at 4°C. Immunoprecipitation was performed using a Co-immunoprecipitation Kit (Pierce, IL, USA), following the manufacturer's protocol, and immunoblots were conducted as described above.
Sub-cellular localization and the BiFC assay
To generate the fluorescent constructs, PCR amplified fragments of the target genes were ligated in frame to the C terminus of the eGFP coding region of pUC18 and expressed under the control of the CaMV 35S promoter. To establish BiFC plasmid constructs, the Pikh-1 full length cDNA or its CC domain sequence were fused with the C terminal fragment of the gene encoding yellow fluorescent protein (YFP) in the pUC-pSPYCE vector [49], while the Pikh-2 CC and the intact and various truncated forms of AvrPik-h were fused with the N terminal fragment of YFP in the same vector. For mislocalization analysis, either the NLS or NES segment were amplified, respectively, from the pMD20T-NLS (the vector harboring an annealed oligo SV40 NLS; [50]) and pMD20T-NES (the vector harboring an annealed oligo PK1 NES; [51]) plasmids with forward and reverse primers containing appropriate restriction sites, and inserted into the GFP-fused or BiFC constructs. Since a monopartite NLS in the DNA-binding domains of the rice auxin response factor ARF19 is responsible for its nuclear localization [52], a truncated element containing the monopartite NLS (ARF19IV) fused to mCherry was prepared in a pMD20T vector (TaKaRa) to generate a mCherry::ARF19IV construct, which was then used to assess nuclear localization. The resulting constructs were used in transient assays based on the polyethylene glycol-mediated transfection of rice protoplasts isolated from ten day old cv. Nipponbare seedlings [53], [54]. After a 30 h incubation at 22°C, the protoplasts were examined by confocal microscopy (Leica TCS SP5), and representative images were shown. Each experiment was repeated at least three times with similar results using independent samples, and at least 30 protoplasts were observed in each independent sample. Nuclear and cytosolic fractions in rice protoplasts expressing various combinations of the BiFC constructs were separated by using a Sub-cellular Fractionation Kit (BestBio, Shanghai, China), following the manufacturer's protocol, and immunoblots were conducted as described above.
Transient expression in N. benthamiana
Pikh-1, Pikh-2 and AvrPik-h fragments were ligated into the binary vectors, pCXSN-FLAG, pCXSN-HA and pCXSN-Myc [48], respectively. To create the redirected Pikh-2 versions, the NLS and NES segments were introduced into the Pikh-2 construct as described above. In addition, the HA-Pikh-2 segment under the control of its native promoter was cloned into the pCAMBIA1300 vector. Cultures of A. tumefaciens strain GV3101 carrying one of these constructs were infiltrated into the leaves of five-week old N. benthamiana plants according to the procedure above-mentioned. The appearance of HR was monitored three days after infiltration. The p35S-INF1 [55] construct was used as a positive control in each experiment.
Transient expression in rice protoplasts
Pikh-1 and Pikh-2 fragments under the control of their respective native promoters, as well as AvrPik-h fragments under the control of the maize ubiquitin promoter, were each inserted into the pMD20T vector. The FLAG-NLS, FLAG-NES, HA-NLS and HA-NES fragments were amplified, respectively, from the constructs created for the transient expression assays, and inserted to the Pikh-1 or Pikh-2 constructs. The resulting constructs, along with a reporter plasmid construct which contained the firefly luciferase gene, were co-transfected into rice protoplasts using a polyethylene glycol-based method [53], [54]. An estimate of the concentration of protoplasts in each transfection reaction was determined using BCA protein assay kit (Pierce). Protoplasts were harvested by centrifugation 40 h after incubation at 22°C, and a luciferase assay conducted using a Luciferase Assay System (Promega). Luciferase activity was set in relation to cell concentration. Each assay consisted of three technical and three biological replicates. All statistical analyses were conducted using SPSS v16.0 software (http://www-01.ibm.com/software/analytics/spss/). For protein expression analysis, PEG-mediated transfections and total protein extraction were performed as described previously [54].
Supporting Information
The genomic location of Pik-h .
(TIFF)
Molecular characterization of Pik-h related transgenic progeny.
(TIFF)
Alignment of predicted polypeptide sequences encoded by the Pik paired genes. (A) Pik-1/Pikm-1/Piks-1/Pikp-1/Pikh-1; (B) Pik-2/Pikm-2/Piks-2/Pikp-2/Pikh-2.
(PPT)
Confirmation of protein expression by Western blot analysis. (A) Yeast two-hybrid assay; (B) Negative control for BiFC assay; (C) N. benthamiana-based transient assay; (D) Rice protoplast-based luciferase assay.
(TIFF)
Sub-cellular localization of the full-length and truncated versions of three proteins involved in Pik-h -mediated resistance. (A) Pikh-1 and a GFP alone control; (B) Pikh-2 and a GFP alone control; (C) AvrPik-h; (D) Mislocation of Pikh-1 and Pikh-2 by using NLS and NES.
(PPT)
Negative controls for BiFC assay.
(TIFF)
Cell fractionation-based Western blot assay.
(TIFF)
Assessment of the autoactivity of AvrPik-hFL, AvrPik-hFL, Pikh-1FL and Pikh-2FL using a rice protoplast-based luciferase assay.
(TIFF)
The relationship between recognition (race) and binding specificity of the Pik and AvrPik alleles. (A–D) The binding specificity of the five Pik allelic proteins: full-length (FL), CC domain, and RAXT1 motif. Three AvrPik alleles (AvrPik-D, -E, and –A; [37]), and Pikh-2CC (FL level only) were selected for binding specificity test via yeast two hybrid system (Clontech, CA). (E) Race specificity of the five Pik alleles. Five monogenic lines for each of the Pik alleles tested with the three AvrPik alleles via general pathotype test.
(TIFF)
Primers, markers and constructs used in the study.
(PDF)
Acknowledgments
We are grateful to Jörg Kudla (University of Münster, Germany) and Dapeng Zhang (Tsinghua University, China) for providing the BiFC vectors, to Guoliang Wang (Ohio State University, USA) for the gift of the Agrobacterium-mediated transformation vectors, to Sophien Kamoun (Sainsbury Laboratory, Norwich, UK) for providing the p35S-INF1 constructs, and to Qianhua Shen (Institute of Genetics and Developmental Biology, Chinese Academy of Science, China) for providing the NES and NLS constructs. We are also indebted to Huishan Guo (Institute of Microbe, CAS, China) for the valuable guidance in co-immunoprecipiation assay. We also thank Dr. Thomas Mitchell (The Ohio State University, USA) for critical reading of the manuscript.
Funding Statement
This research was carried out with financial support from the National Natural Science Foundation (U1131003; 30870102;31170247) and the National 973 Project (2011CB1007001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Heath M (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334. [DOI] [PubMed] [Google Scholar]
- 2. Flor H (1971) Current status of the gene-for-gene concept. Annu Rev Phytopathol 9: 275–296. [Google Scholar]
- 3. Meyers B, Kozik A, Griego A, Kuang H, Michelmore R (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis . Plant Cell 15: 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, et al. (2009) RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J 60: 218–226. [DOI] [PubMed] [Google Scholar]
- 5. Nombela G, Williamson V, Muñiz M (2003) The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci . Mol Plant-Microbe Interact 16: 645–649. [DOI] [PubMed] [Google Scholar]
- 6. Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, et al. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AvrPia and Avr1-CO39 by direct binding. Plant Cell 25: 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dangl J, Jones J (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- 8. Chen Y, Liu Z, Halterman D (2012) Molecular determinants of resistance activation and suppression by Phytophthora infestans effector IPI-O. PLoS Pathogens 8: e1002595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dodds P, Lawrence G, Catanzariti A, Teh T, Wang C, et al. (2006) Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA 103: 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yahiaoui N, Brunner S, Keller B (2006) Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J 47: 85–98. [DOI] [PubMed] [Google Scholar]
- 11. Takken F, Tameling W (2009) To nibble at plant resistance proteins. Science 324: 744–746. [DOI] [PubMed] [Google Scholar]
- 12. Takken F, Albrecht M, Tameling W (2006) Resistance proteins: Molecular switches of plant defence. Curr Opin Plant Biol 9: 383–390. [DOI] [PubMed] [Google Scholar]
- 13. Frost D, Way H, Howles P, Luck J, Manners J, et al. (2004) Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol Plant-Microbe Interact 17: 224–232. [DOI] [PubMed] [Google Scholar]
- 14. Krasileve K, Dahlbeck D, Staskawicz B (2010) Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate Oomycete effector. Plant Cell 22: 2444–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ade J, DeYoung B, Golstein C, Innes R (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Caplan J, Mamillapalli P, Burch-Smith T, Czymmek K, Dinesh-Kumar S (2008) Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132: 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bernoux M, Ve T, Williams S, Warren C, Hatters D, et al. (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling and autoregulation. Cell Host Microbe 9: 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maekawa T, Cheng W, Spiridon L, Töller A, Lukasik E, et al. (2011) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9: 187–199. [DOI] [PubMed] [Google Scholar]
- 19. Burch-Smith T, Schiff M, Caplan J, Tsao J, Czymmek K, et al. (2007) A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen Q, Saijo Y, Mauch S, Biskup C, Bieri S, et al. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103. [DOI] [PubMed] [Google Scholar]
- 21. Wirthmueller L, Zhang Y, Jones J, Parker J (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol 17: 2023–2029. [DOI] [PubMed] [Google Scholar]
- 22. Cheng Y, Germain H, Wiermer M, Bi D, Xu F, et al. (2009) Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis . Plant Cell 21: 2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mang H, Qian W, Zhu Y, Qian J, Kang H, et al. (2012) Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing muclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis . Plant Cell 24: 1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Slootweg E, Roosien J, Spiridon L, Petrescu A, Tameling W, et al. (2010) Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tameling W, Nooijen C, Ludwig N, Boter M, Slootweg E, et al. (2010) RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22: 4176–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao Z, Chung E, Eitas T, Dangl J (2011) Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci USA 108: 7619–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eitas T, Dangl J (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13: 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sinapidou E, Williams K, Nott L, Bahkt S, Tor M, et al. (2004) Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis . Plant J 38: 898–909. [DOI] [PubMed] [Google Scholar]
- 29. Peart J, Mestre P, Lu R, Malcui I, Baulcombe D (2005) NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol 15: 968–973. [DOI] [PubMed] [Google Scholar]
- 30. Loutre C, Wicke T, Travella S, Galli P, Scofield S, et al. (2009) Two different CC-NBS-LRR genes are required for Lr10-mediated leaf rust resistance in tetraploid and hexaploid wheat. Plant J 60: 1043–1054. [DOI] [PubMed] [Google Scholar]
- 31. Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, et al. (2008) Two adjacent nucleotide-binding site-leucine rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180: 2267–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S, Song M, Seo Y, Kim H, Ko S, et al. (2009) Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two CC-NB-LRR genes. Genetics 181: 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, et al. (2011) A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J 66: 467–479. [DOI] [PubMed] [Google Scholar]
- 34. Yuan B, Zhai C, Wang W, Zeng X, Xu X, et al. (2011) The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor Appl Genet 122: 1017–1028. [DOI] [PubMed] [Google Scholar]
- 35. Zhai C, Lin F, Dong Z, He X, Yuan B, et al. (2011) The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol 189: 321–334. [DOI] [PubMed] [Google Scholar]
- 36. Kiyosawa S (1982) Genetics and epidemiological modeling of breakdown of plant disease resistance. Ann Rev Phytopathol 20: 93–117. [Google Scholar]
- 37. Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A, et al. (2012) Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J 76: 894–907. [DOI] [PubMed] [Google Scholar]
- 38. Tameling W, Elzinga S, Darmin P, Vossen J, Takken F, et al. (2002) The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14: 2929–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rairdan G, Collier S, Sacco M, Baldwin T, Boettrich T, et al. (2008) The coiled-coil and nucleotide binding domains of the potato RX disease resistance protein function in pathogen recognition and signaling. Plant Cell 20: 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jia Y, McAdams S, Bryan G, Hershey H, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19: 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DeYoung B, Innes R (2006) Plant NBS-LRR proteins in pathogen sensing and host defense. Nature Immunology 7: 1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deslandes L, Olivier J, Peeters N, Feng D, Khounlotham M, et al. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Catanzariti A, Dodds P, Ve T, Kobe B, Ellis J, et al. (2010) The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol Plant-Microbe Interact 23: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Caplan J, Padmanabhan M, Dinesh-Kumar S (2008) Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 3: 126–135. [DOI] [PubMed] [Google Scholar]
- 45. Michelmore R, Paran I, Kesseli R (1991) Identification of marker linked to disease resistance gene by bulk segregant analysis: a rapid mathod to detect markers in specific genomic regions using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ryu H, Han M, Lee S, Cho J, Ryoo N, et al. (2006) A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep 25: 836–8247. [DOI] [PubMed] [Google Scholar]
- 47. Yoshida K, Saitoh H, Fujisawa S, Kanzaki H, Matsumura H, et al. (2009) Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae . Plant Cell 21: 1573–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen S, Songkumarn P, Liu J, Wang G (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walter M, Chaban C, Schütze K, Bastistic O, Weckermann K, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438. [DOI] [PubMed] [Google Scholar]
- 50. Haasen D, Köhler C, Neuhaus G, Merkle T (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucinerich nuclear export signals in Arabidopsis thaliana. Plant J 20: 695–705. [DOI] [PubMed] [Google Scholar]
- 51. Wen W, Meinkoth J, Tsien R, Taylor S (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473. [DOI] [PubMed] [Google Scholar]
- 52. Shen C, Wang S, Huang Y, Wu Y, Zhang S, et al. (2010) Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.) J Exp Bot. 61: 3971–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen S, Tao L, Zeng L, Vega-Sanchez M, Umemura K, et al. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–27. [DOI] [PubMed] [Google Scholar]
- 54. Shen C, Zhang Q, Li J, Bi F, Yao N (2010) Induction of programmed cell death in Arabidopsis and rice by single-wall carbon nanotubes. American J Bot 97: 1602–1609. [DOI] [PubMed] [Google Scholar]
- 55. Huitema E, Vleeshouwers V, Cakir C, Kamoun S, Govers F (2005) Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INF1, INF2A, and INF2B of Phytophthora infestans . Mol Plant-Microbe Interact 18: 183–193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The genomic location of Pik-h .
(TIFF)
Molecular characterization of Pik-h related transgenic progeny.
(TIFF)
Alignment of predicted polypeptide sequences encoded by the Pik paired genes. (A) Pik-1/Pikm-1/Piks-1/Pikp-1/Pikh-1; (B) Pik-2/Pikm-2/Piks-2/Pikp-2/Pikh-2.
(PPT)
Confirmation of protein expression by Western blot analysis. (A) Yeast two-hybrid assay; (B) Negative control for BiFC assay; (C) N. benthamiana-based transient assay; (D) Rice protoplast-based luciferase assay.
(TIFF)
Sub-cellular localization of the full-length and truncated versions of three proteins involved in Pik-h -mediated resistance. (A) Pikh-1 and a GFP alone control; (B) Pikh-2 and a GFP alone control; (C) AvrPik-h; (D) Mislocation of Pikh-1 and Pikh-2 by using NLS and NES.
(PPT)
Negative controls for BiFC assay.
(TIFF)
Cell fractionation-based Western blot assay.
(TIFF)
Assessment of the autoactivity of AvrPik-hFL, AvrPik-hFL, Pikh-1FL and Pikh-2FL using a rice protoplast-based luciferase assay.
(TIFF)
The relationship between recognition (race) and binding specificity of the Pik and AvrPik alleles. (A–D) The binding specificity of the five Pik allelic proteins: full-length (FL), CC domain, and RAXT1 motif. Three AvrPik alleles (AvrPik-D, -E, and –A; [37]), and Pikh-2CC (FL level only) were selected for binding specificity test via yeast two hybrid system (Clontech, CA). (E) Race specificity of the five Pik alleles. Five monogenic lines for each of the Pik alleles tested with the three AvrPik alleles via general pathotype test.
(TIFF)
Primers, markers and constructs used in the study.
(PDF)