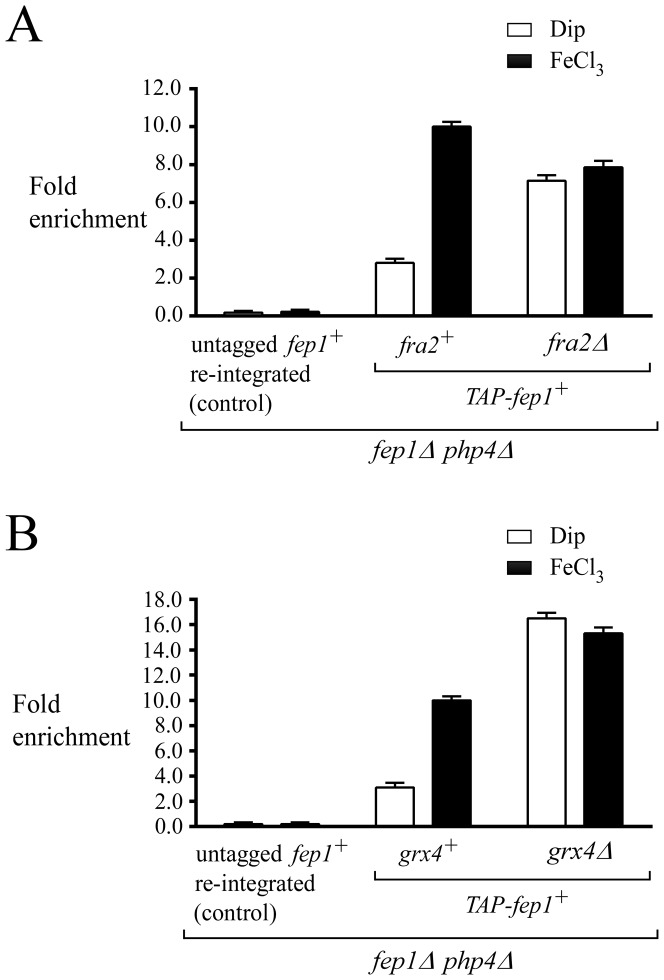
Figure 4. Iron starvation-dependent dissociation of Fep1 from its target gene promoter fio1+ requires Fra2.
A, ChIP analysis of the fio1+ promoter in a fep1Δ php4Δ double mutant or a fep1Δ php4Δ fra2Δ triple mutant strain harboring an integrated untagged or TAP-tagged fep1+ allele. Cells were grown to logarithmic phase in the presence of FeCl3 (100 µM), and then incubated in the presence of 250 µM Dip or 250 µM FeCl3 (Fe) for 30 min. Chromatin was immunoprecipitated using an anti-mouse IgG antibodies and a specific region of the fio1+ promoter was analyzed by qPCR to determine Fep1 occupancy. Binding of TAP-Fep1 to the fio1+ promoter was calculated as the enrichment of a specific fio1+ promoter region relative to a 18S ribosomal DNA coding region in which no GATA box was present. ChIP data were calculated as values of the largest amount of chromatin measured (fold enrichment). Results are shown as the average +/− standard deviation of a minimum of three independent experiments. B, ChIP analysis was performed on the fio1+ promoter in fep1Δ php4Δ or fep1Δ php4Δ grx4Δ cells expressing an untagged or TAP-tagged fep1+ allele. This analysis was performed as a control experiment because it is known that deletion of the grx4+ gene leads to constitutive promoter occupancy by Fep1. ChIP data were calculated and presented as described in panel A.