Figure 2.
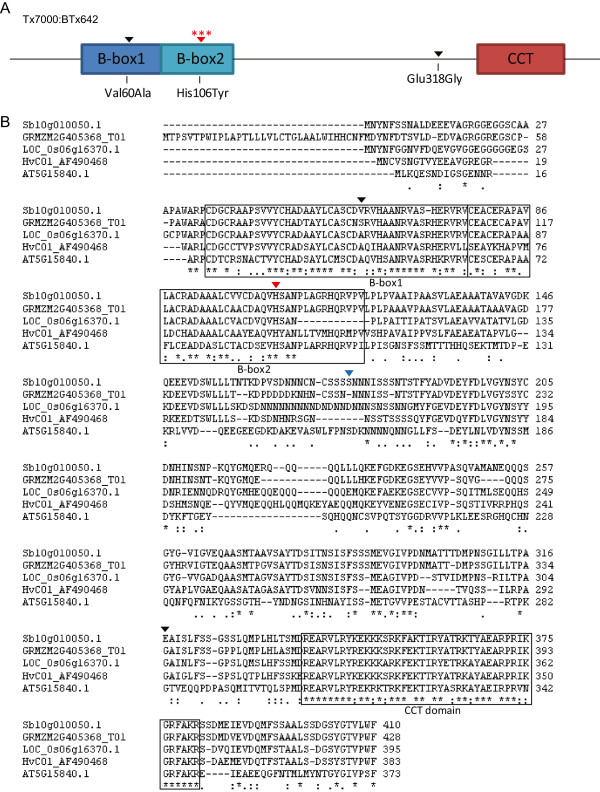
Multiple alignment analysis of CONSTANS homologs. A. Protein structure of SbCO showing domains characteristic of CONSTANS-like gene families: B-box1, B-box2 and CCT domain are boxed. Red asterisks above the His106Tyr mutation indicate that this functional mutation was also identified in rice and Arabidopsis. B. Multiple sequence alignments of CO homologs from sorghum (Sb10g010050, SbCO), maize (GRMZM2G405368_T01, conz1), rice (Os06g16370, OsHd1), barley (AF490468, HvCO1) and Arabidopsis (AT5G15850, AtCO). The sorghum sequence used for alignment was derived from BTx623 (SbCO-1). Protein residues conserved among all 5 species are underscored by asterisks. Amino acid residues underscored by a colon indicate residues of strongly conserved properties, while residues underscored by a period indicate residues with more weakly similar properties. One amino acid substitution distinguishes BTx623 (SbCO-1) and Tx7000 (SbCO-2) (marked with blue arrow). Unique amino acid substitutions that distinguish BTx623 and BTx642 (Sbco-3) are marked with black arrows (tolerant) and a red arrow (intolerant).
