Figure 2.
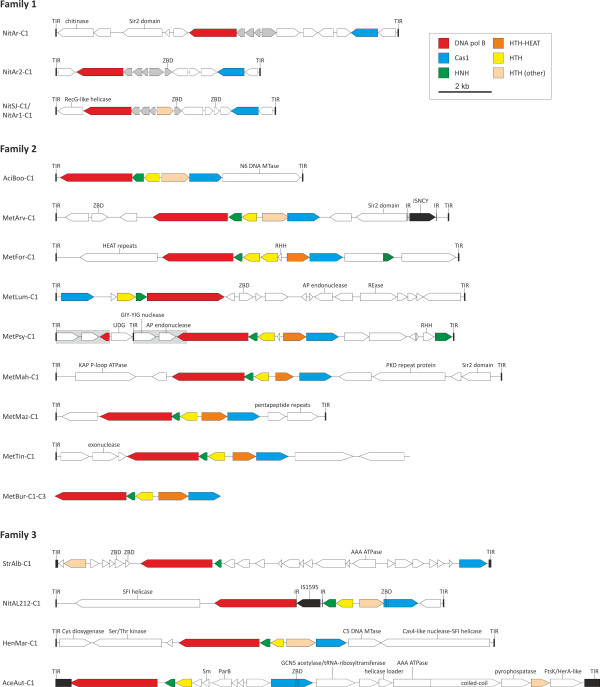
Casposon genome maps. The three families of casposons are indicated. The precise nucleotide coordinates of depicted casposons can be found in Additional file 1: Table S1. Predicted protein-coding genes are indicated with arrows, indicating the direction of transcription. The color key for the designation of the common genes is shown in the top right area of the figure. ‘HTH (other)’ denotes proteins that are not orthologous but nevertheless contain HTH domains. Terminal inverted repeats (TIR) are shown with black rectangles and their sequences are shown in Additional file 1: Figure S2. The grey boxes outlined with a broken line in MetPsy-C1 depict duplicated regions. The striped green arrows represent genes encoding divergent HNH proteins. Abbreviations: ZBD, zinc-binding domain-containing protein; HNH, HNH family endonuclease; HTH, helix-turn-helix proteins; MTase, methyltransferase; RHH, ribbon-helix-helix protein; REase, restriction endonuclease; AP, apurinic/apyrimidinic; UDG, uracil-DNA glycosylase; SFI, superfamily I; IR, inverted repeat.
