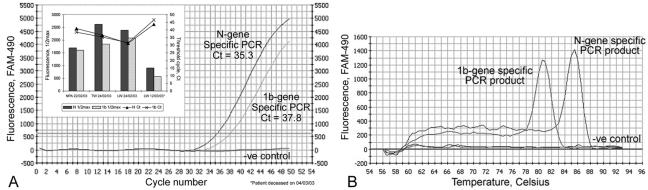
FIG. 5.
Amplification curve (A) and melting curve (B) of real-time quantitative PCR specific to 1b and N gene of SARS CoV. (A) Amplification plot of fluorescence intensity versus the number of PCR cycles. One-microliter samples of cDNAs from NPA, tracheal dispersion, and lung biopsy samples from patients with clinical symptoms were used as templates for PCRs. Fifty cycles of PCR were performed to achieve the saturation phase of the reaction. The x axis indicates the cycle number of the quantitative PCR, while the y axis represents the fluorescence intensity (FAM-490) above the background signal. The horizontal gray line indicates the threshold value calculated by the maximum curvature approach, and the baseline cycle CT was calculated automatically. The inset shows half-maximal fluorescence values (1/2max) and CT values of both PCRs with cDNAs from various tissues isolated from a key patient (patient A [1]) at three different time points. TW, tracheal wash; LW, lung wash. (B) Melting curves of PCR products. Melting curve analysis was performed after a 10-min further extension step. The Tm rose from 56 to 94°C for 76 0.5-s steps, while each set-point temperature was held for 7 s for data collection and analysis. The Tms of the 1b- and N-gene-specific PCR products were 80.5 and 85.5°C, respectively. The x axis indicates the temperature in degrees Celsius, while the y axis represents the fluorescence intensity (FAM-490) above the background signal. One microliter of water was used as a no-template control in the reaction.