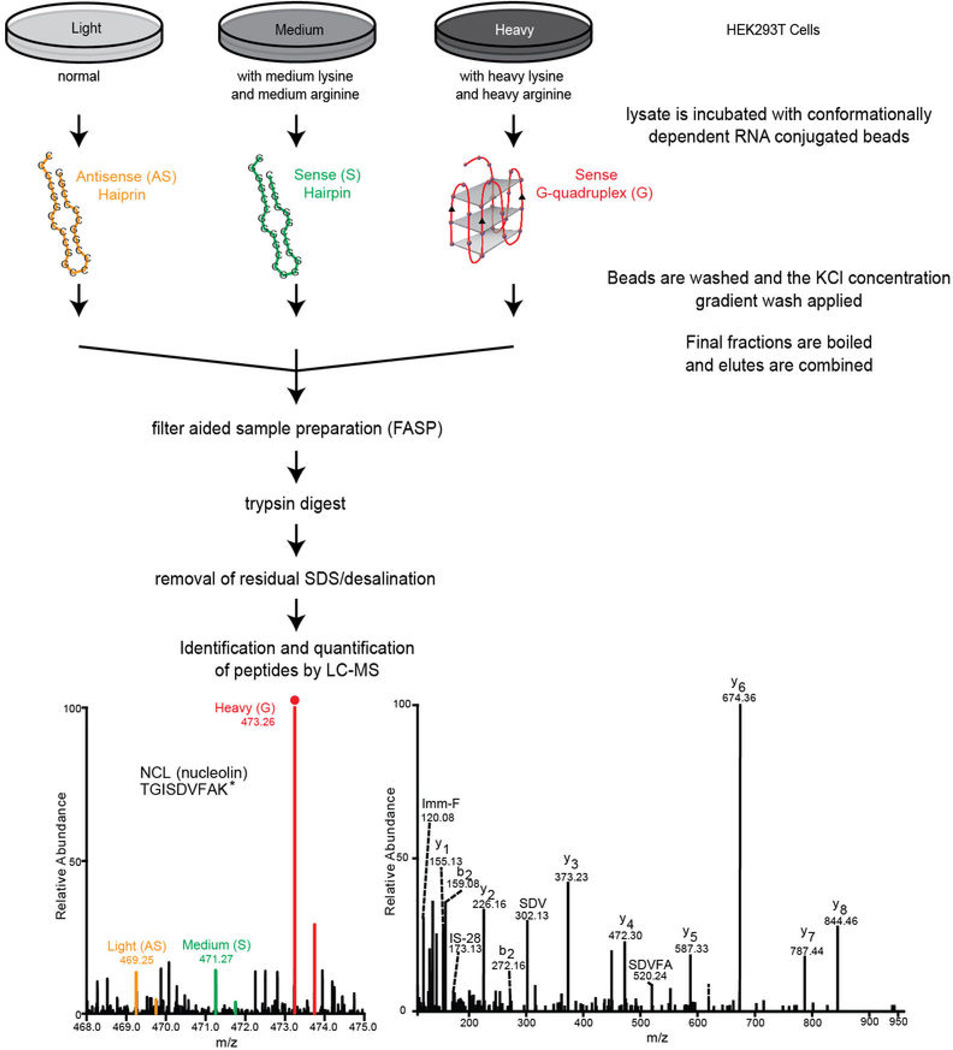
Extended Data Figure 5. Workflow for the SILAC methodology used for identification and quantification of proteins that bind the hexanucleotide repeats in a structurally dependent manner.
HEK293T cells that had complete incorporation of either heavy-, medium-, or lightlabeled isotopes were incubated with either the (GGGGCC)4 G-quadruplex, the (GGGGCC)4 hairpin, or the (CCCCGG)4 hairpin, respectively. The samples were then processed as described in Methods and presented in Figures 3. The final eluted samples were combined and prepared for trypsin digestion using filter-aided sample preparation (FASP). The buffer was exchanged to remove salts and SDS. The sample was then subject to LC-MS/MS analysis as described in the Methods.