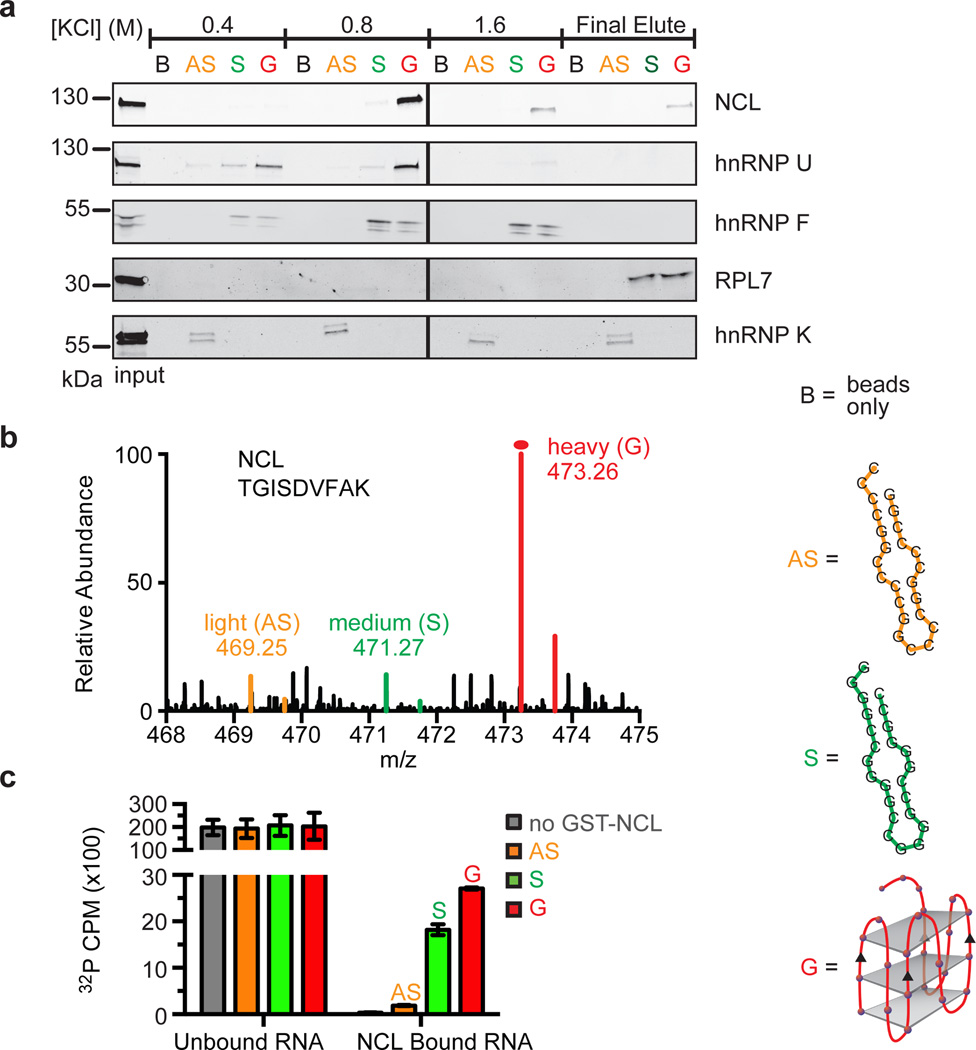
Figure 3. Identification of conformation-dependent C9orf72 HRE RNA-binding proteins.
a) Western blotting analyses of the sequential fractions eluted from a biotinylated-RNA pulldown of HEK293T cells with increasing KCl concentrations. The RNA-binding proteins identified using LC-MS were differentiated among those that recognize different structural motifs, including the RNA antisense (AS) hairpin, (CCCCGG)4; the RNA sense (S) hairpin (GGGGCC)4; and the sense G-quadruplex (G). NCL and hnRNP U have binding preferences for the G motif of the sense RNA. hnRNP F and RPL7 bind both guanine-rich sense sequences, regardless of the underlying RNA structure. hnRNP K prefers binding to the cytosine-rich AS. b) A representative spectrum from SILAC analysis is shown for the preferential binding of NCL to the G motif formed by (GGGGCC)4 compared to the hairpin motifs formed by the sense or antisense sequences. c) An RNA pulldown performed with GST-NCL demonstrates that NCL directly binds (GGGGCC)4 RNA in vitro with highest affinity for the G-quadruplex. Data are means ± s.d. n = 3.