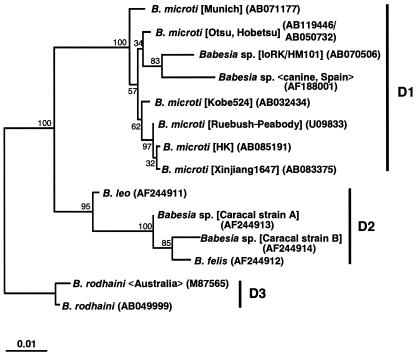
FIG. 1.
Phylogenetic tree based on the SSU rDNA sequences of 14 Babesia spp. closely related to B. microti. Maximum-likelihood analysis (HKY85 + Γ model with eight categories for site rates) reconstructed three monophyletic subgroups, D1, D2, and D3. The sequence of IoRK/HM101 was the closest relative of a Spanish canine isolate in subgroup D1. GenBank accession numbers are given in parentheses; the name of the isolate, strain, clone, or genotype is shown in brackets; and the host origin or the isolation site is shown in chevrons. The SSU rDNA sequences used for the primary analysis are as follows; they were selected on the basis of the results of a FASTA similarity search (http://www.ddbj.nig.ac.jp/E-mail/homology-j.html) (12, 15) by using the SSU rDNA sequence of IoRK/HM101 as a query: B. microti (AB032434, AB119446, AB050732, AB071177, AB083375, AB085191, and U09833), Babesia sp. (AF188001), B. felis (AF244912), B. leo (AF244911), Cytauxzoon felis (L19080), Babesia sp. (AF244913 and AF244914), B. rodhaini (M87565 and AB049999), B. bigemina (X59604), B. bovis (L19077), B. caballi (Z15104), B. canis (L19079), B. divergens (U07885), B. equi (Z15105), B. gibsoni (AF175300 and AF158702), B. odocoilei (U16369), Babesia sp. (AB053216), Piroplasmida gen. sp. CA1 (AF158703), Babesia sp. WA1 (AY027815), Theileria parva (AF013418), Theileria mutans (AF078815), Plasmodium falciparum (M19172), Plasmodium berghei (M14599), Toxoplasma gondii (U03070), Hepatozoon sp. Boiga (AF297085), Sarcocystis tenella (L24383), and Eimeria tenella Houghton (AF026388).