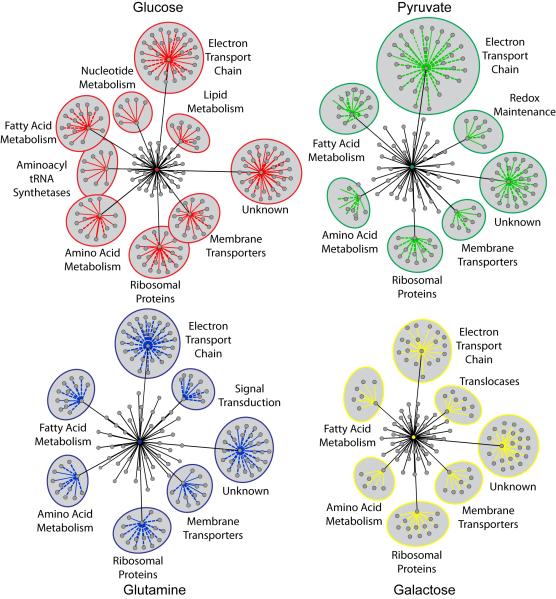
Figure 2. Mitochondrial Functions Impacting Cellular Bioenergetics.
Cytoscape (see Methods section) was used to define mitochondrial functions affecting ATP levels in response to each metabolic condition. Central colored nodes represent individual nutrient sources. Gray nodes radiating from the central node represent all mitochondrial functions associated with siRNAs altering ATP by ≥25%. The distance between each functional node and the central node is proportional to the number of genes associated with that function that increased or decreased ATP by ≥ 25% in that condition. Functions with five or more associated genes altering ATP by ≥ 25% are labeled and include the associated genes, represented as nodes with colored edges. Solid colored edges represent increased ATP levels; dotted edges represent decreased ATP levels. The distance between each gene node and the associated functional node is proportional to the magnitude of ATP level change. See also Table S3 and Figure S2.