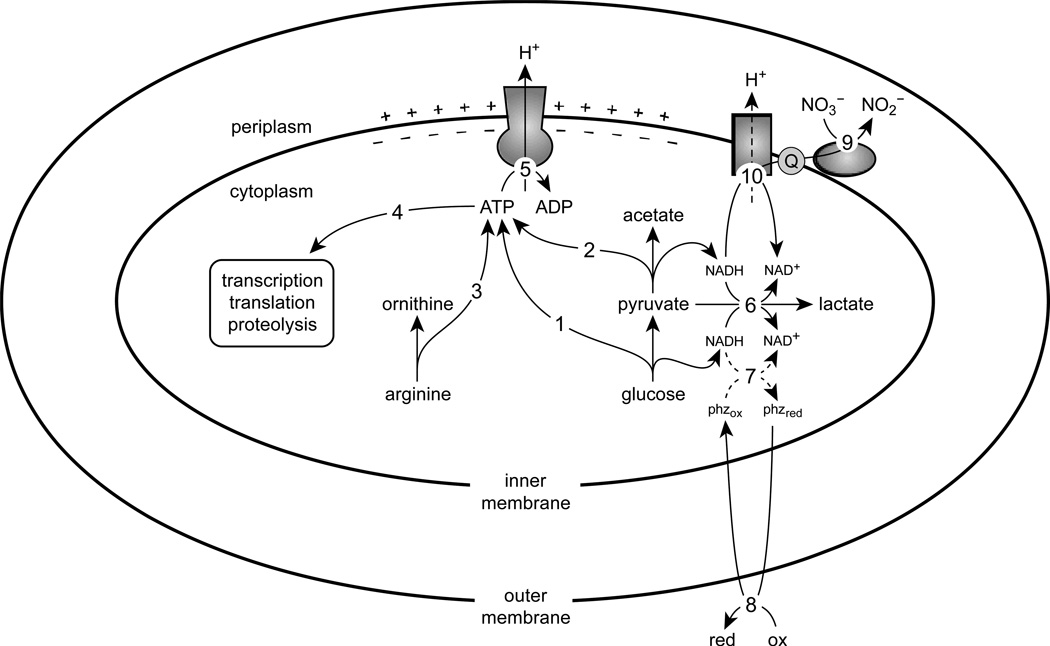
Figure 7.
A metabolic model of survival in P. aeruginosa. The ATP synthesized from glycolysis (1), pyruvate fermentation (2), and the arginine deiminase pathway (3) is used for essential processes including transcription, translation, and proteolysis (4). The F1FO-ATPase complex hydrolyses ATP to translocate protons across the inner membrane (5), thereby generating a proton-motive force. Excess reducing equivalents from pyruvate oxidation can be dispensed by converting pyruvate to lactate (6). Phenazines can also regenerate oxidants for metabolism (7); although NAD(P)H is a possible electron donor for phenazines in vivo (Price-Whelan et al., 2007), the dashed lines indicate that the nature of this interaction has not been elucidated. Phenazines are then oxidized extracellularly (8) and can be reused. Nitrate reductase can reduce nitrate to nitrite using electrons from the quinone pool (9), permitting oxidation of NADH via NADH dehydrogenase (10), which can result in proton translocation depending on which dehydrogenase complex is used.