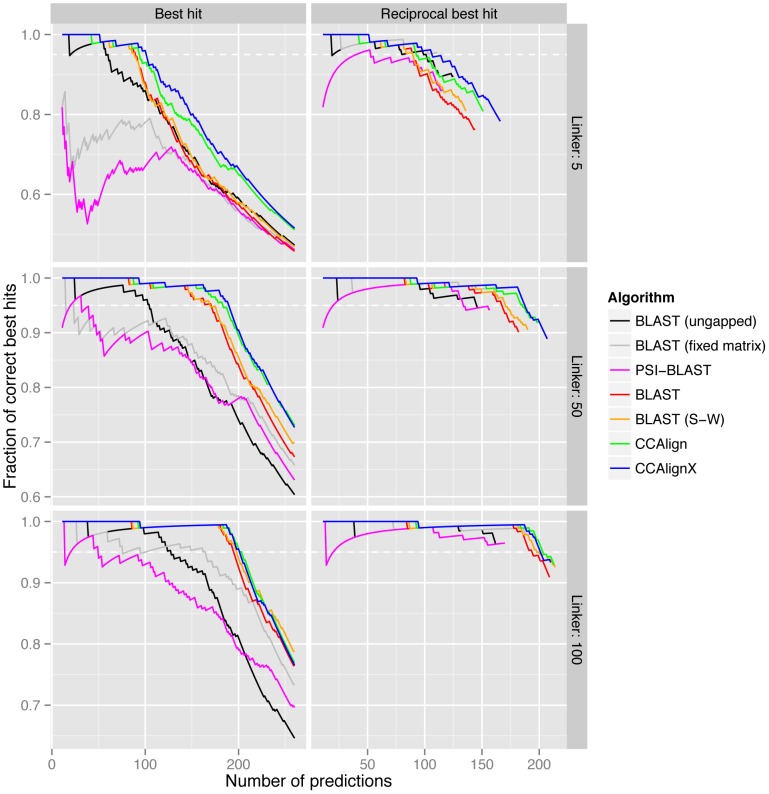
Figure 2. Benchmarking of algorithms.
Alignment algorithms were applied to coiled-coil proteins from a set of manually annotated yeast orthologs. For each protein, it was then determined if the best hit was actually annotated as orthologous. Non-coiled-coil parts of the proteins were reduced to respective linker lengths (shown on the right of the panels) to increase the difficulty of detecting the true homolog. CCAlign and CCAlignX, the two algorithms that take the sequence properties of coiled-coils into account, perform better than standard algorithms. Dashed horizontal lines indicate a 5% false discovery rate. For clarity, data for less than ten predictions is omitted. (S-W: Smith-Waterman algorithm).