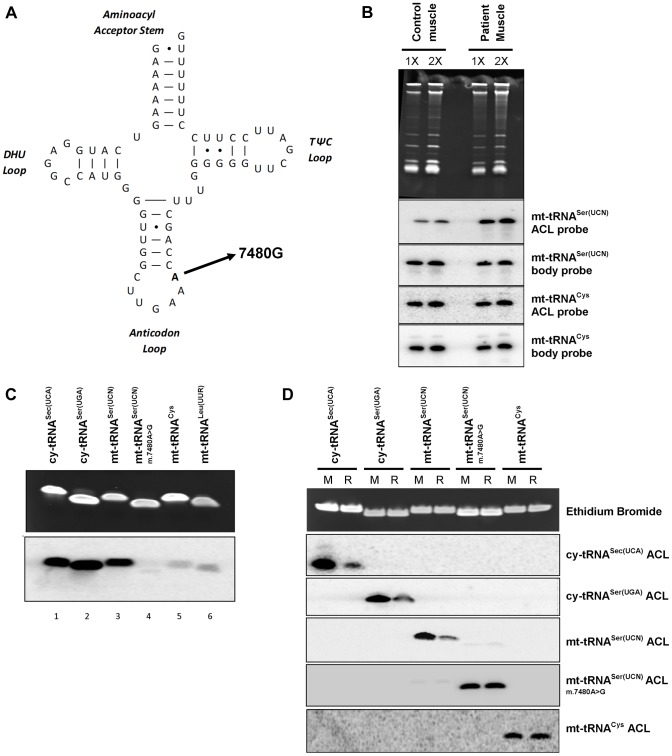
Figure 6. A mt-tRNASer(UCN) point mutation, m.7480A>G, impairs i6A37 modification.
A) The previously reported m.7480A>G mutation [15] is located at position 38 in the anticodon loop (ACL) of mt-tRNASer(UCN), in the A36A37A38 recognition sequence of TRIT1. B) The i6A modification of position 37 is decreased (stronger binding of the mt-tRNASer(UCN) double ACL probe) by ∼86% in patient skeletal muscle relative to control muscle. Binding of the ACL probe to the non-TRIT1 substrate, mt-tRNACys, confirmed even loading. The steady-state level of mt-tRNASer(UCN) in patient skeletal muscle is 30% lower than in control muscle (calculated using the two body probes). Both control and patient samples were run in duplicate using 2 µg (1X) and 4 µg (2X) of RNA as indicated above the lanes. C) In vitro isopentylation assay using purified TRIT1, 14C-DMAPP, and synthetic minihelixes representing the anticodon stem loops (ASLs) of the tRNAs indicated above the lanes. The upper blot is an ethidium bromide-stained gel of the ASLs after in vitro reaction indicating even loading, whilst the lower blot is the autoradiograph obtained after 3 days of exposure. D) The PHA6 assay was validated and the loss of i6A37 modification in m.7480A>G mutant mt-tRNASer(UCN) confirmed by in vitro modification of synthetic templates. For reaction samples (R), synthetic RNA minihelixes were in vitro modified using unlabeled-DMAPP and recombinant His-TRIT1. For mock-treated samples (M), all the components except the His-TRIT1 enzyme were added. After purification, the RNA samples were transferred to a membrane which was repeatedly hybridized, stripped and rehybridized with 5 different 32P-labeled ASL oligo probes as indicated to the right. Ethidium bromide staining of the gel confirmed equal loading of each pair of reactions (mock and reaction samples) for each synthetic ACL interrogated.