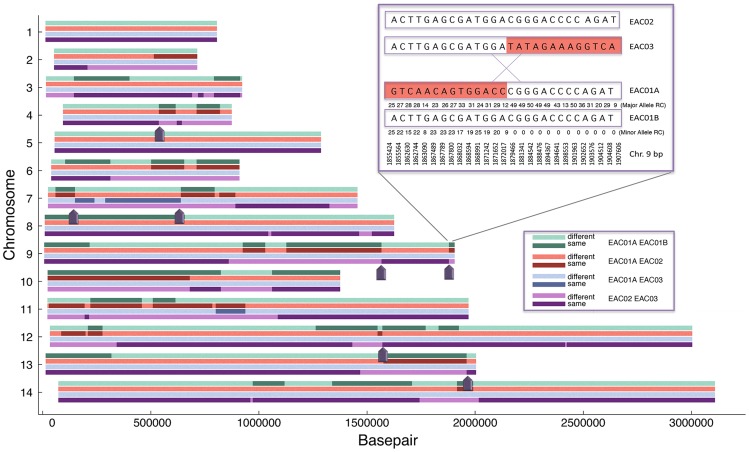
Figure 4. Multiple two-way comparisons show evidence of reciprocal recombination events.
To ensure that only high quality positions would be used, the 55,399 genotyped loci were stringently filtered. The filtering criteria was that there had to be more than 20 read counts for the major allele of each comparison (EAC01, EAC02, and EAC03), and that all three strains' SNV positions differed from the reference. A likelihood function was generated examining 10,000 basepair windows for recombination and assumed that each transition (same to different in each pairwise comparison taken individually) could be considered a chance of recombination. A smoothing kernel was applied to differentiate if a position was likely to be the same by chance or due to recombination and depended on the distance of identical neighboring SNVs (the further separated identical SNVs were, the chances of a true recombination event was much less). Only edges with Z scores greater than 4.188 (99th percentile) were taken as true edges. For the inset, the numbers below the line represent read count. On the right side of the recombination event, there is no minor allele in EAC01 and thus EAC01A and EAC01B are presumed to be identical, while on the left side, the minor allele (EAC01B) read count is relatively high. RC = Read Count. Dark arrows indicate reciprocal recombination events found in three or more comparisons.