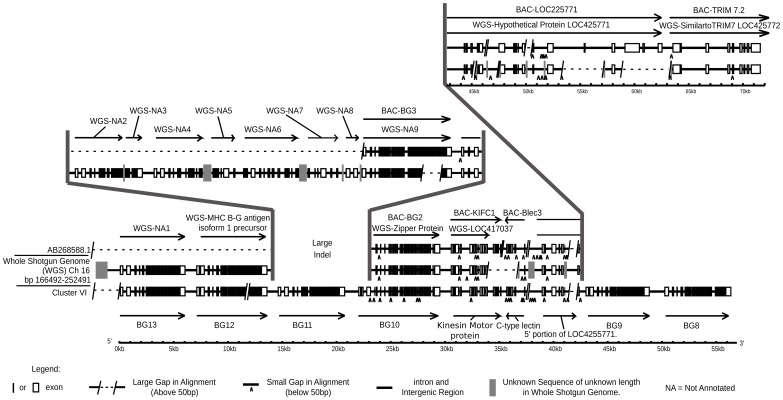
Figure 9. Comparison of cosmid cluster VI from the B12 haplotype with the BQ haplotype from a red junglefowl, showing regions of virtual identity separated by two large indels, one in the middle of the sequences and the other where the red junglefowl haplotype (but not the B12 haplotype) continues into the TRIM region.
Genomic organisation on bottom line is from cluster VI of this paper (accession number KC955130) compared to two sequences from the BQ haplotype, middle line from the WGS sequence assembly (nucleotides 166492–252491 on chromosome 16) and top line from the sequence of a BAC from the same individual chicken (accession number AB268588.1). Note that there exist differences between the WGS and BAC sequences, and further that the WGS assembly has regions of unknown sequence with only approximate length. WGS-NA indicates genes not annotated by ENSEMBL at the time of this analysis.