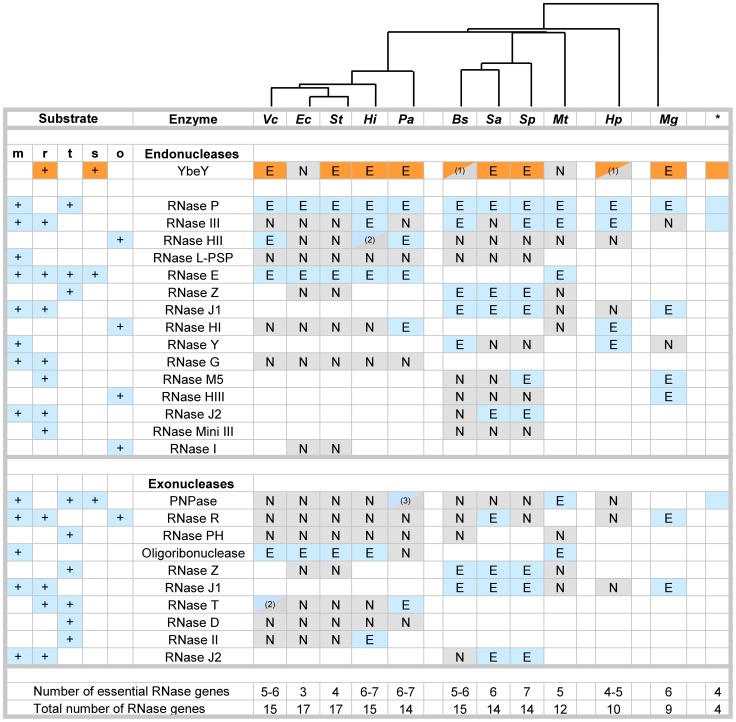
Figure 1. Essential RNases in human pathogens.
Compilation of major RNases in V. cholerae (Vc), E. coli (Ec), S. typhi (St), H. influenzae (Hi), P. aeruginosa (Pa), B. subtilis (Bs), S. aureus (Sa), S. pneumoniae (Sp), M. tuberculosis (Mt), H. pylori (Hp), and M. genitalium (Mg). The asterisk indicates RNases present in the minimal bacterial genome set [15]. RNases with dual endo- and exonuclease functions are listed twice, but are counted only once to assess the total number of genes. Blank squares indicate that no homolog for the respective RNase could be identified. The designation of RNases as putatively essential (E) and non-essential (N) is based on the availability of deletion or transposon mutants according to Materials and Methods. (1) Current databases list these RNases as non-essential; follow-up experiments suggest these RNases could by essential (personal communication; BW Davies, LA Simmons, Y Furuta). (2) RNases were listed as essential in one database and non-essential in a second database. (3) Only mutants with transposon insertions in the last 10% of the gene are available. The key roles of RNases in mRNA (m), rRNA (r), tRNA (t) and sRNA (s) metabolism are indicated; other functions (o). The phylogenetic tree on top is based on alignment of various YbeY protein sequences using Geneious software.