Figure 1.
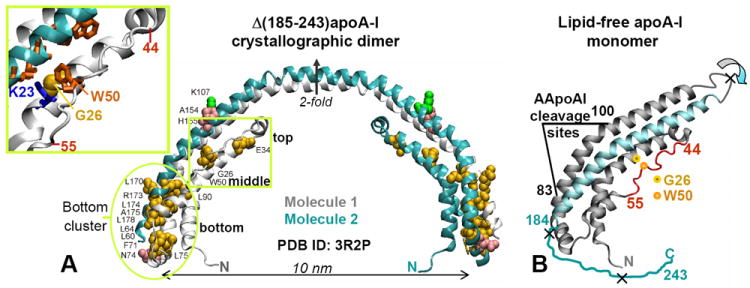
Locations of the AApoAI mutations in the structure of free apoA-I. (A) X-ray crystal structure of human lipid-free Δ(185-243)apoA-I (PDB ID 3R2P) solved by Mei and Atkinson [10] with the sites of all known AApoAI mutations mapped [38]. The structure is a crystallographic dimer stabilized by two symmetry-related four-helix bundles. Dimer-forming molecule 1 (gray) and 2 (teal) and the crystallographic two-fold axis relating them are shown. Residues that are mutated in AApoAI are shown by spheres: point mutations (yellow), deletions (green), and frame shifts (pink). Oval encircles the “bottom” half of the helix bundle where most of the AApoAI point mutations are located. Boxed region contains the sites of all known AApoAI point mutations located in the middle (G26R and W50R) or near the top (E34K) of the helix bundle. Insert shows zoomed-in structure in this boxed region; side chains of G26, W50 and K23 are shown. (B) Proposed structure of lipid-free apoA-I monomer obtained from the dimer structure via the domain swapping (indicated by circular arrows) of residues 121-184 (in teal) from molecule 2 to molecule 1 around the 2-fold axis [10]. Thin irregular line represents C-terminal residues 185-243 that are largely unfolded in solution and have been deleted from Δ(185-243)apoA-I to facilitate crystallization [10]. Extended segment in residues 44-55 is in red. The cleavage sites between residues 83 and 100, which are characteristic of the protein fragments found in patient-derived AApoAI deposits, are indicated; X indicate major cleavage sites upon processing of free apoA-I by the broad-specificity proteases. Locations of the two mutations studied in this work, G26R and W50R, are shown.
