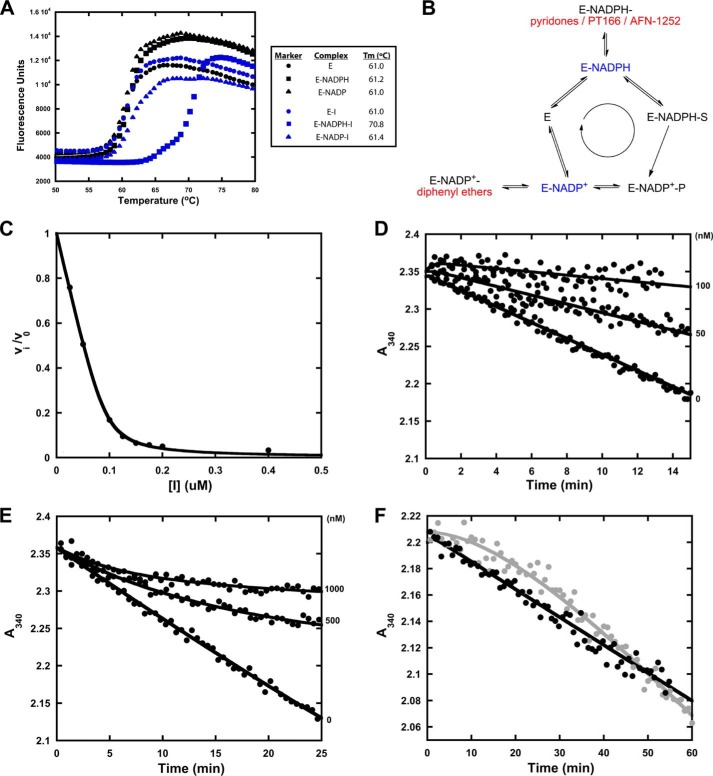
FIGURE 2.
Different mechanisms of saFabI inhibition. A, thermal shift analyses of saFabI bound to NADPH, NADP+, and/or inhibitor (CG400549). The measurement variability is approximately ±0.2 °C. B, distinct mechanisms of saFabI inhibition. Based on our recent kinetic and structural results, we reasoned that the enzyme binds NADPH first followed by the substrate (31). In contrast to diphenyl ethers (highlighted in red), which bind to the enzyme-product complex generated via catalysis (E-NADP+, depicted in blue) (31), pyridone compounds (A), PT166 (supplemental Fig. S2) and AFN-1252 (71) (all shown in red), preferentially inhibit saFabI at the enzyme-substrate complex state (E-NADPH, depicted in blue). C, representative plot of fractional velocity (v/v0) as a function of inhibitor concentration for a potent pyridone (CG400549, in this example). The shape is characteristic of tight-binding inhibition. The best fit curve to the Morrison quadratic equation (Equation 3) yields Kiapp = 4.73 ± 0.50 nm and [E]T = 92.73 ± 2.52 nm (R2 = 0.99). D, representative set of forward progress curves with pyridone-based inhibitors of saFabI. The plot depicts rapid-onset inhibition at different PT173 concentrations. E, as a reference, this plot illustrates the slow-onset inhibition of saFabI by the diphenyl ether PT04 (31). Note the clear observation of curvature that is absent in D. F, jump dilution curve for CG400549 (black) following preincubation with NADPH and saFabI. The jump dilution curve for the slow off diphenyl ether inhibitor PT52 (gray) following preincubation with NADP+ and saFabI is shown as reference (full recovery of activity; tR = 30 min, where tR is the residence time) (31). The lack of curvature for CG400549 is consistent with rapid off kinetics.