FIGURE 5.
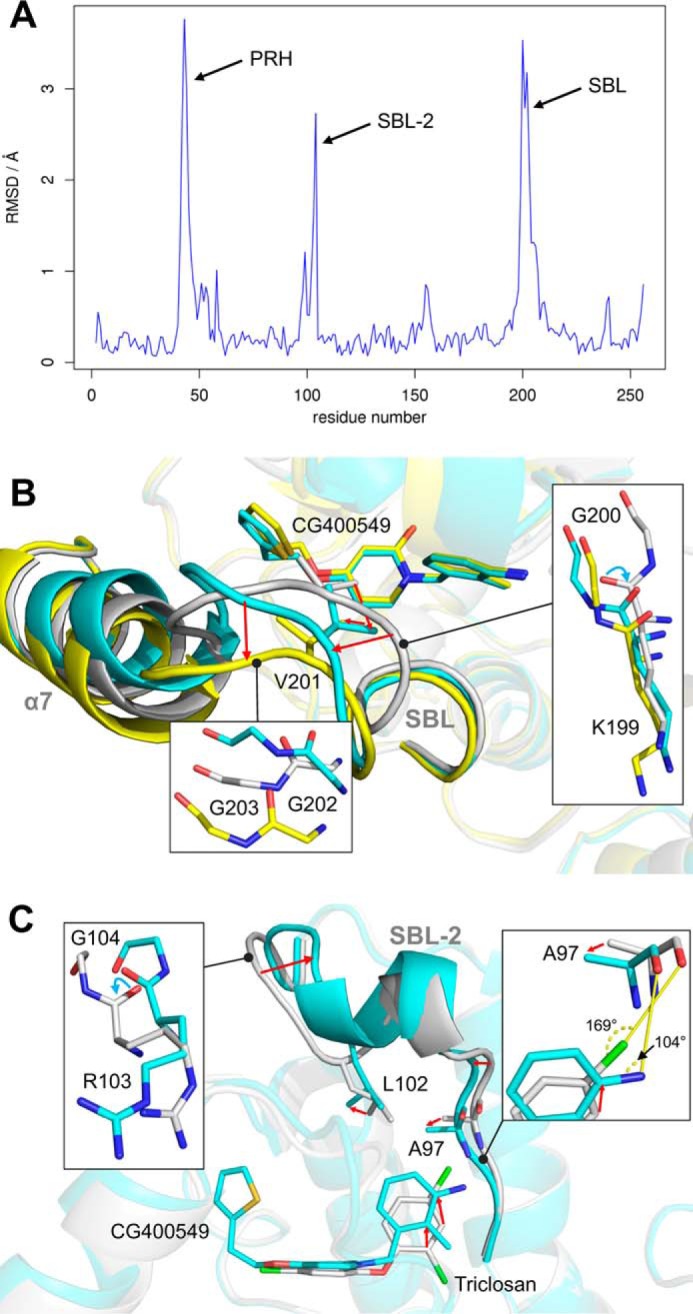
Structural variations between pyridone and diphenyl ether ternary complex structures. A, structural differences between diphenyl ether and pyridone ternary complexes. Per residue root mean square deviation (RMSD) values between our triclosan-bound (PDB code 4ALI, subunit H) and CG400549-bound (CG400549-I, subunit C) structures were calculated using Theseus (79) and are plotted against the residue number. The inhibitor-bound structures of these two scaffolds differ considerably in three regions of the protein (PRH = phosphate recognition helix α2; SBL-2 = substrate-binding loop 2; SBL = substrate-binding loop). B, conformational states of the SBL. The different subunits of our CG400549-I structures reveal two distinct states of the SBL and the attached helix α7 (shown in cyan and yellow for subunits A and C, respectively; residual parts of the protein are displayed in transparent colors for clarity). Compared with these conformations, the SBL is more closed in our triclosan-bound structure (shown in gray; PDB code 4ALI, subunit H). Detailed views of the three different conformations are displayed in the insets. Red arrows indicate the conformational changes from the closed to the open substrate-binding loop states. Cyan arrows highlight backbone flips. C, conformational states of the SBL-2. Selected residues, the inhibitors, and the SBL-2 are shown for the CG400549-I (cyan; subunit A) and triclosan-bound structures (gray; PDB code 4ALI, subunit H).
