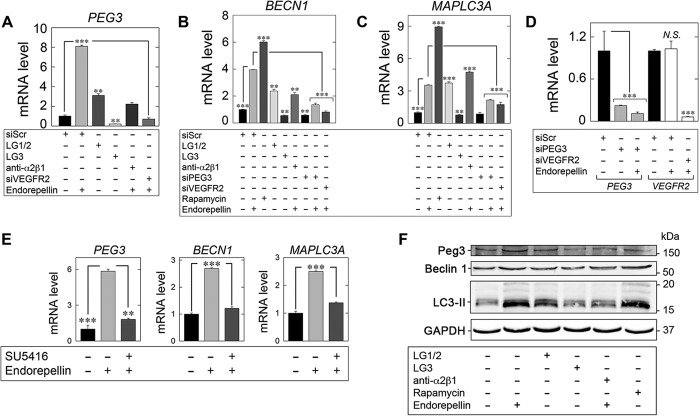
FIGURE 4.
Endorepellin regulates an autophagy transcriptional program via PEG3 and VEGFR2. A–C, expression of autophagy genes in HUVEC via qPCR under various experimental conditions involving LG1/2 (150 nm, 6 h), LG3 (150 nm, 6 h), and pre-incubation with the α2β1 blocking mAb (10 μg/ml, 30 min) in the presence of endorepellin (200 nm, 6 h) or via RNAi-mediated silencing of PEG3 (siPEG3, 80 pm) alone or in combination with endorepellin (200 nm, 6 h) or in the absence of VEGFR2 (siVEGFR2, 80 pm) following stimulation with endorepellin (200 nm, 6 h) relative to siScr (20 pm) controls for B, PEG3, C, BECN1, or D, MAPLC3A. Rapamycin (40 nm, 2 h) served as a positive control. D, verification of PEG3 (siPEG3, 80 pm) and VEGFR2 (siVEGFR2, 80 pm) RNAi-mediated knockdown relative to siScramble (siScr, 20 pm) controls in the presence or absence of endorepellin (200 nm, 6 h) via quantitative real time-PCR analysis in HUVEC. N.S., not significant. E, qPCR analysis of PEG3, BECN1, and MAPLC3A following endorepellin alone (200 nm, 6 h) or via pre-incubation (30 min) with the VEGFR2 small tyrosine kinase inhibitor SU5416 (30 μm) followed by treatment with endorepellin (200 nm, 6 h) in HUVEC. F, representative immunoblot of Peg3, Beclin 1, and LC3-II following incubation with either endorepellin (200 nm, 6 h), LG1/2 (150 nm, 6 h), or LG3 (150 nm, 6 h). The blocking mAb anti-α2β1 (10 μg/ml) was pre-incubated for 30 min prior to endorepellin stimulation (200 nm, 6 h). Rapamycin (40 nm, 2 h) served as a positive control. All gene expression changes were first normalized to the endogenous housekeeping gene, ACTB, calculated via the ΔΔCt method and reported as fold changes ±S.E. For immunoblotting, GAPDH served as a loading control. All data are representative of three independent trials run in quadruplicate replicates. **, p < 0.01; ***, p < 0.001.