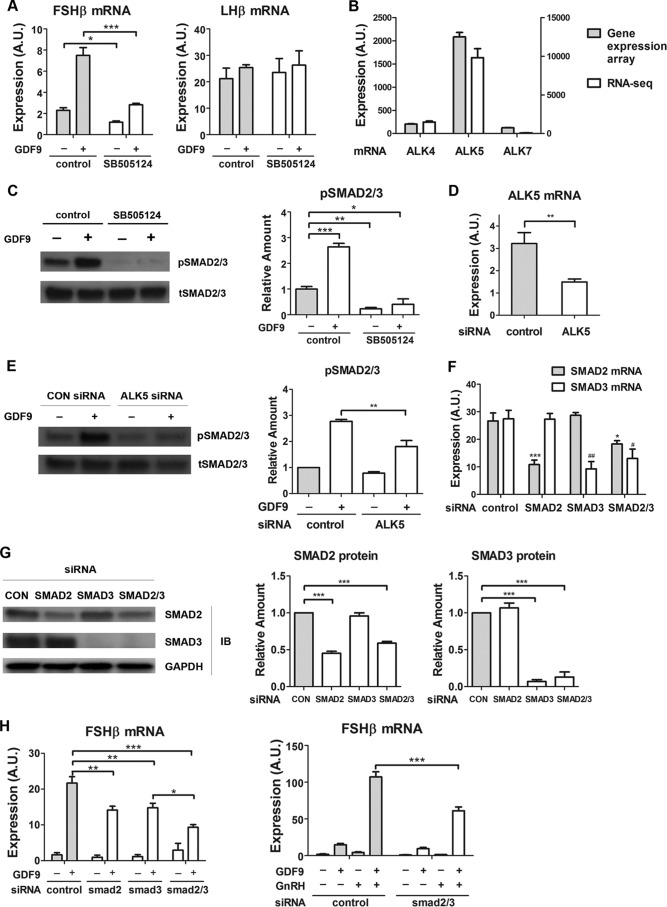
FIGURE 5.
Involvement of ALK5 and Smad2/3 in GDF9 activation of FSHβ gene expression. A, LβT2 cells were serum-starved overnight, pretreated with SB-505124 (100 nm) or control (0.01% dimethyl sulfoxide) for 30 min, and stimulated with either GDF9 (100 ng/ml) or vehicle for 6 h. Two-way ANOVA with Bonferroni post test corrections was used. ***, p < 0.001; **, p < 0.01; *, p < 0.05. B, LβT2 cells were transfected with scrambled siRNA for 48 h. RNA samples were subjected to whole genome expression profiling analysis (left y axis) and to RNA-Seq assays (right y axis), as described under “Experimental Procedures.” C, cells were serum-starved overnight, pretreated with SB-505124 (100 nm) or control (0.01% dimethyl sulfoxide) for 30 min, and stimulated with either GDF9 (100 ng/ml) or vehicle for 1 h. Left, whole cell lysates were subjected to a quantitative Western blot analysis using total Smad2/3 (tSMAD2/3)- and phospho-Smad2/3 (pSMAD2/3)-specific antibodies. Right, quantification of Western blot densitometry from three independent experiments is plotted as mean ± S.E. (error bars). One-way ANOVA was used. n = 3; *, p < 0.05. D, cells were transfected with either scrambled (control) or ALK5 siRNA. Two days after transfection, cells were harvested for qPCR measurement of ALK5 mRNA. E, cells were transfected with either scrambled (CON) or ALK5 siRNA on day 1. On day 2, cells were serum-starved overnight. On day 3, cells were stimulated with either GDF9 (100 ng/ml) or vehicle for 1 h. Left, whole cell lysates were subjected to a quantitative Western blot analysis using total Smad2/3 (tSMAD2/3)- and phospho-Smad2/3 (pSMAD2/3)-specific antibodies. Right, quantification of Western blot densitometry from three independent experiments is plotted as mean ± S.E. (error bars). Two-way ANOVA was used. n = 3; *, p < 0.05. F and G, knockdown efficiencies of Smad2, Smad3, and Smad2/3 siRNAs in LβT2 cells. LβT2 cells were transfected with either scrambled or Smad2/3 siRNA. F, 2 days after transfection, cells were harvested for qPCR measurement of Smad2 and Smad3 mRNA. One-way ANOVA with Bonferroni post test corrections was used. G, left, 2 days after transfection, whole cell lysates were subjected to a quantitative Western blot (IB) analysis using Smad2-, Smad3-, and Smad2/3-specific antibodies. Middle and right, quantification of Western blot densitometry from three independent experiments, plotted as mean ± S.E. (error bars). One-way ANOVA was used. n = 3; *, p < 0.05. H, left, LβT2 cells were transfected with either scrambled or Smad2/3 siRNA for 48 h, serum-starved overnight, and stimulated with either GDF9 (100 ng/ml) or vehicle for 6 h. Right, LβT2 cells were transfected with either scrambled or Smad2/3 siRNA for 48 h, serum-starved overnight, and stimulated with either GDF9 (100 ng/ml) for 6 h, vehicle, GnRH (1 nm) for 2 h (followed by 4 h without GnRH), or both GDF9 and GnRH. Two-way ANOVA with Bonferroni post test corrections was used. ***, p < 0.001; **, p < 0.01; *, p < 0.05. A.U., arbitrary units.