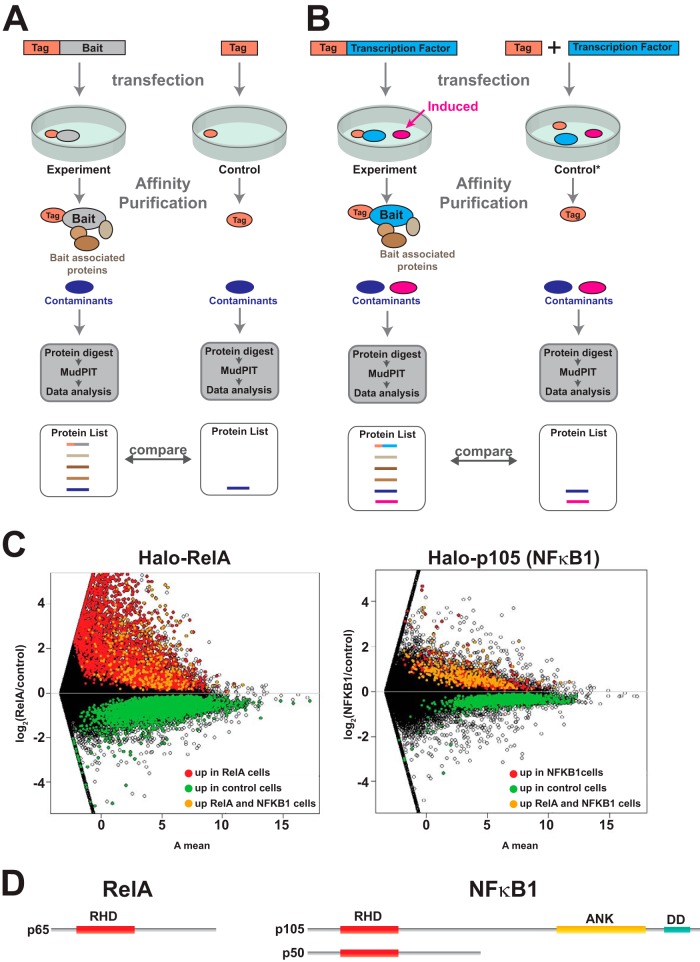
Fig. 2.
Ectopically expressed bait affects global gene expression. A, comparing proteins purified from experimental, affinity-tagged bait transfected cells with affinity-tag-only control transfected cells assuming that the transfections do not differ in their effects on global patterns of gene expression. In this model, the same contaminants (shown in dark blue) co-purify nonspecifically in both bait and control experiments and are accounted for during data processing. B, Alternatively transfecting cells with DNA encoding the bait might cause significant changes in global gene expression (for example, if the bait is a transcription factor). It is possible that a protein induced or up-regulated following transfection with such bait might co-purify nonspecifically as a contaminant (pink). In this case a different control might be needed (control denoted by asterisk). C, changes in global gene expression patterns among three biological replicates of cells transfected with the Halo-control plasmid and three replicates of cells transfected with either the transcription factor Halo-RelA or the NFκB family member Halo-p105 (NFκB1). Changes in gene expression are represented using an MA plot (intensity ratio versus average intensity). Differentially expressed genes are shown in color (genes called by the Cuffdiff algorithm with an adjusted p value < 0.05 and fold change (FC) > 1.4) (16). D, the DNA binding transcription factor RelA contains a Rel homology domain (RHD). In addition to the RHD, p105 (NFκB1) contains ankyrin repeats (ANK) and a death domain (DD). The p105 protein is partially processed into the NFκB1 transcription factor p50.