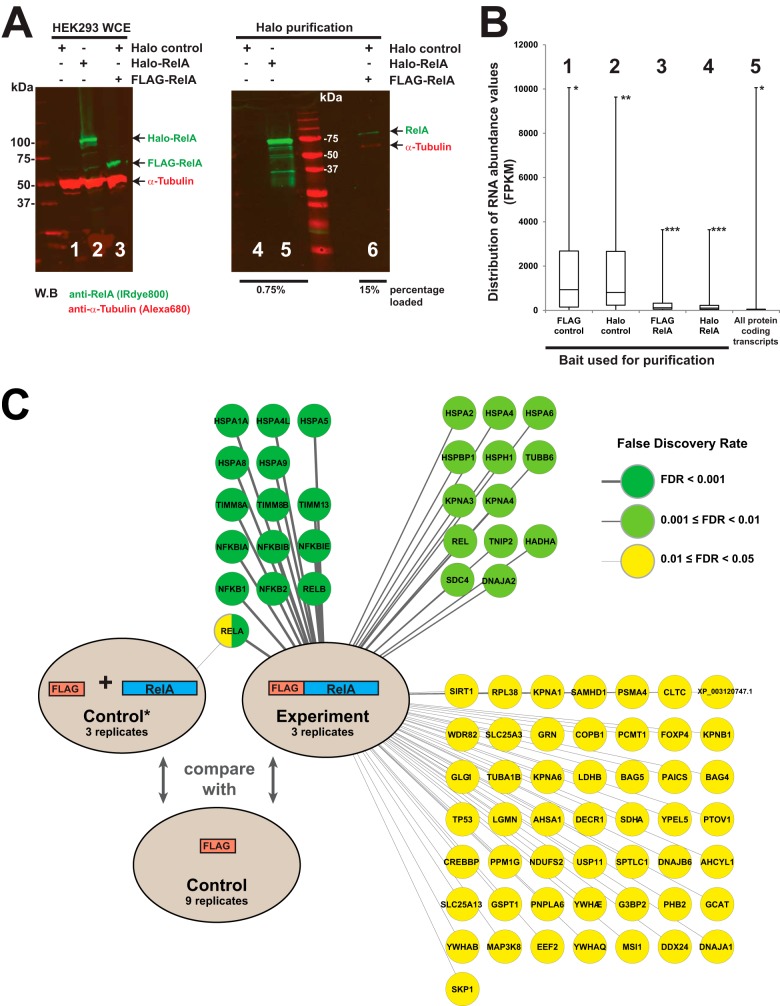
Fig. 3.
Small amounts of highly expressed proteins are purified nonspecifically during affinity purification. A, small but detectable amounts of recombinant overexpressed FLAG-RelA are observed after enrichment of proteins by Halo purification. HEK293T cells were transfected with DNA plasmids expressing the Halo tag alone, Halo-RelA, or FLAG-RelA, and the resulting whole cell extracts were subjected to Halo affinity purification as described in the text. Proteins present in the eluates were fractionated via SDS-PAGE and visualized through Western blotting. RelA protein was detected using anti-RelA rabbit polyclonal primary antibodies and IRDye™-800-labeled goat anti-rabbit IgG secondary antibodies (green); α-tubulin was detected using anti-α-tubulin mouse monoclonal primary antibodies and Alexa Fluor 680–labeled anti-mouse IgG secondary antibodies (red). A Li-Cor Odyssey infrared imaging system was used to detect the fluorescently labeled secondary antibodies. B, the distribution of RNA transcript abundances for different groups of proteins identified via mass spectrometry (groups 1–4, supplemental Data S4) or for all cellular proteins (group 5). FKPM values used to assess mRNA abundance were derived from RNA purified from cells transfected with either Halo-control (groups 1, 2, and 5) or Halo-RelA plasmids (groups 3 and 4). Box plots of the distribution of FKPM values indicate sample minimum, lower quartile, median, upper quartile, and sample maximum value. Groups of proteins analyzed are the top 50 detected in the control purifications via MudPIT (groups 1 and 2), the top 50 enriched in the experimental RelA purifications (groups 3 and 4), and all protein-coding transcripts in Halo-control transfected cells (group 5). FKPM values for the overexpressed RelA baits were not calculated for groups 3 and 4. Proteins in each group with the highest FKPM values were *the ribosomal protein RPL14 (groups 1 and 5), **the ribosomal protein RPS3 (group 2), and ***the NFκB inhibitor NFKBIA (groups 3 and 4). C, small amounts of contaminating, overexpressed Halo-RelA, but no other proteins, were identified via MudPIT/PLGEM analysis (FDR cutoff of 0.05) in FLAG purified Halo-RelA-expressing cells. In contrast, 77 bait-associated proteins were identified in FLAG-RelA-expressing cells subject to FLAG immunoaffinity purification (FDR cutoff of 0.05) (supplemental Data S1 and S5).