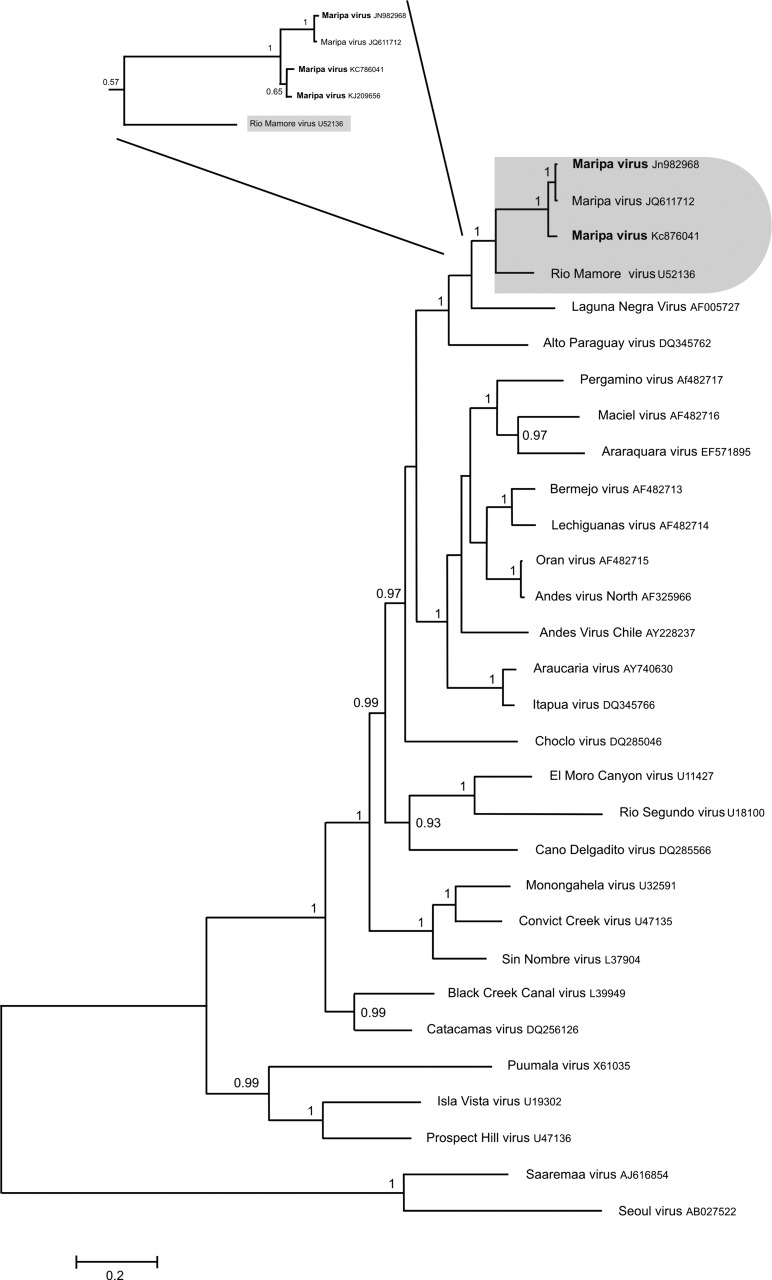
Figure 1.
Phylogenetic tree constructed using Bayesian methods (1,000,000 replicates; Mr. Bayes, version 3.1b) based on 1,308 bp of the S segment of 30 hantaviruses. The tree is based on the GTR + I + G model of nucleotide evolution. The virus names are associated with their accession numbers. The Maripa virus sequences generated in this study are in bold. The association between Maripa viruses and Rio Mamoré virus is labeled. Support for nodes is provided by the posterior probabilities of the corresponding clades. All resolved nodes have posterior probability greater than 0.8. Scale bar indicates nucleotide sequence divergence among hantavirus sequences. Focus corresponds to the phylogenetic tree based on 345 bp of the S segment of the hantavirus detected in Oligoryzomys captured around the four human cases.