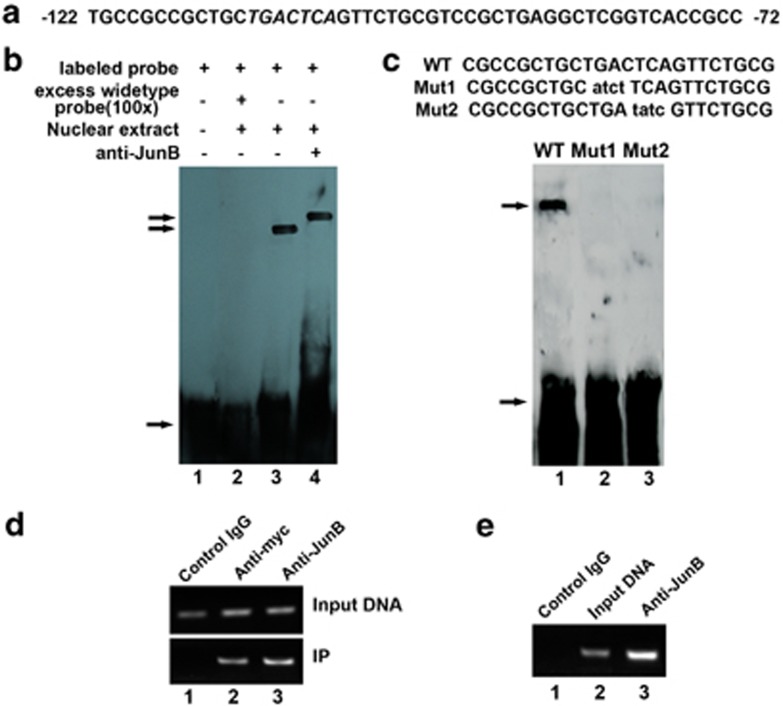
Figure 3.
JunB binds to the AP1 sequence of the IRE1a promoter in vitro (EMSA) and in vivo (ChIP). (a) DNA sequence of the 50-bp AP1 (−122 to −72 bp) of the IRE1a promoter. Putative JunB-binding elements (AP1) are underlined. (b) JunB binds to the AP1 of the IRE1a promoter in vitro (EMSA). Ten micrograms of nuclear extracts (NE) prepared from C2C12 cells transfected with pcDNA3.1(−)-JunB was incubated with Dig-labeled JunB-binding site (ER stress response element) probe in reaction buffer (20 μl). For competition experiments, a 100-fold excess of WT oligodeoxynucleotide was added. After 15 min of incubation, the Dig-labeled AP1 probe was added, and the reaction mixture was incubated for an additional 15 min and analyzed by gel electrophoresis. Arrows indicate free DNA probe (bottom) and DNA/protein complex (up). (c) Mutations of AP1 sequence abolish the binding of JunB to the IRE1a gene promoter (DNA). EMSA was performed using the same protein fractions as in b. Probes are as indicated above the lanes. The specific protein–DNA band is indicated by an arrow. Mutant nucleotides are lowercase and underlined. The binding intensity of JunB to these probes is indicated as follows:+, binding; −, no binding. (d) Myc-tagged JunB binds to the transfected IRE1a-specific reporter construct −2065IRE1aluc. C2C12 cells transfected with −2065IRE1aluc and the expression plasmid pcDNA3.1(−)-JunB were cross-linked by formaldehyde treatment and lysed. Cell lysates were subjected to immunoprecipitation with control IgG (lane 1) or with anti-myc (lane 2) or anti-JunB (lane 3) antibody. Purified DNA from the cell lysate (Input DNA; upper panel) and DNA recovered from immunoprecipitation (IP; lower panel) were amplified by PCR. (e) Endogenous JunB binds to the AP1 sequence of the IRE1a gene. ChIP assays were performed in C2C12 cells treated with BMP2 for 3 days. C2C12 cells treated with formaldehyde were lysed, and DNA was sheared by sonication. Cell lysates were subjected to immunoprecipitation with either control IgG (IgG; lane 1) or anti-JunB antibodies (lane 3). DNA recovered from the immunoprecipitation was amplified by PCR. Input DNA (lane 2) was used as a positive control