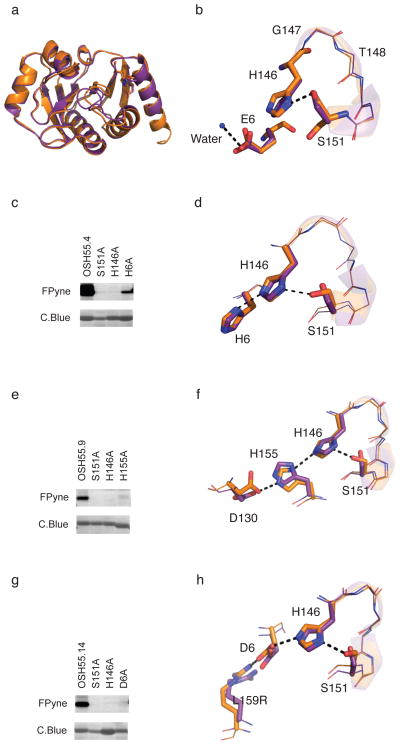
Figure 2. Characterization of OSH55 derived designs.
(a) Structural superposition of design (brown) and crystal structure (purple) of OSH55 with overall backbone RMSD of 0.6 Å. The all-atom RMSD over the binding pocket residues is 0.34 Å. (b) A zoom-in view of the active site of OSH55 showing the orientation of Ser151 and His146 as designed and Glu6 coordinating a water molecule. (c,e,g) Selective fluorophosphonate probe labeling of OSH55.4 (c), OSH55.9 (e) and OSH55.14 (g) but not the active-site knockout mutants. Serine reactivity was assessed by gel-based ABPP (top, in-gel fluorescence; bottom, coomassie staining, see Supplementary Fig. 16 for full-size gel images. The experiments were performed in duplicates with consistent results). There is much less labeling in the catalytic triad knockouts but no decrease in the amount of protein. (d,f,h) Superimposition of the designed catalytic triad active sites (brown) overlaid on the experimentally determined crystal structures (purple) for OSH55.4 (d), OSH55.9 (f) and OSH55.14 (h); the designed hydrogen bond networks are recapitulated with very high accuracy.