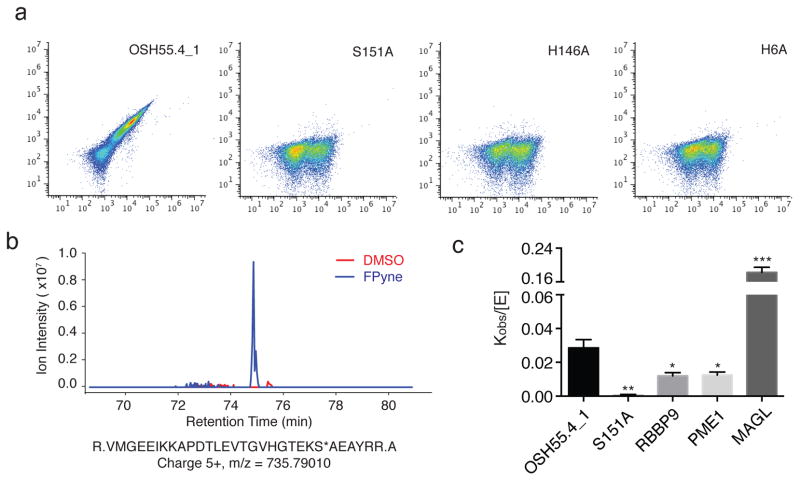
Figure 3. Organophosphate reactivity of OSH55.4_1 is comparable to native enzymes.
(a) Flow cytometry analysis of OSH55.4_1 and active site knockouts displayed on yeast following incubation with FP-biotin. Signal along x-axis indicates extent of yeast surface display; signal along y-axis; extent of probe binding. The active site residue knockouts abolish binding to FP-biotin. (b) MS1 chromatographic traces of the OSH55.4_1 active-site peptide adducted with FPyne probe. LC-MS/MS experiments (See Methods) identified a fully tryptic peptide (m/z = 735.79010 and z = 5+) with probe modification on the catalytic Ser151 (denoted with an asterisk). The extracted MS1 chromatographic traces (±15 p.p.m.) showed a distinct peak present only in the FPyne-treated (blue) but not in the DMSO-treated (red) sample. (c) Comparison of rates of FP-Rh labeling of the OSH55.4_1 design to those of a representative set of native serine hydrolases by fluorescence polarization. The kobs/[E] (mean ± s.e.m.) value for OSH55.4_1 is greater than those of RBBP9 and PME1. The S151A knockout mutation reduces the reaction rate to near background level (*p<0.05, **p<0.01, ***p<0.001, t-test, comparison with OSH55.4_1).