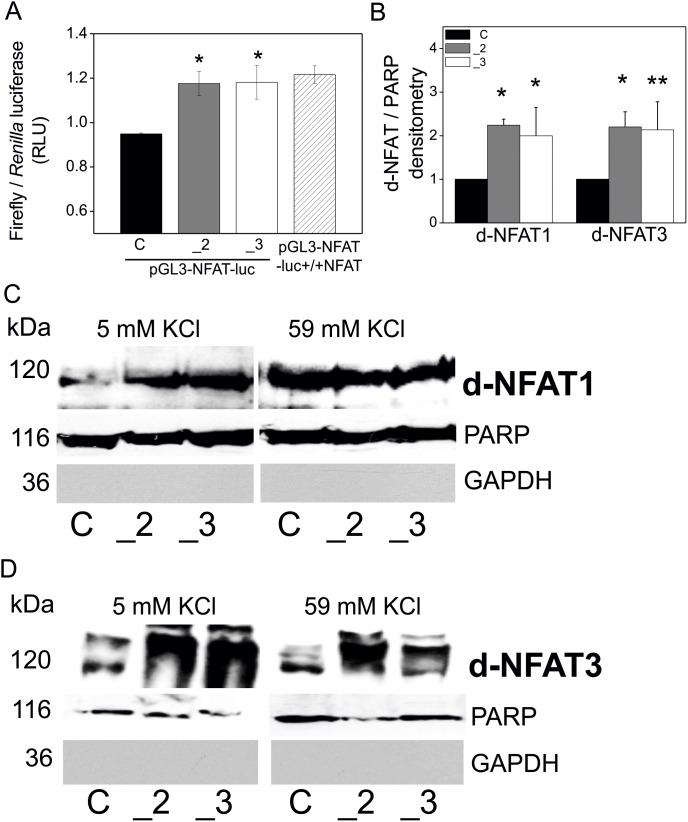
Figure 1. NFAT activation in PC12 cells with reduce PMCAs content.
PC12 cells were transfected with plasmids encoding firefly luciferase under NFAT-dependent promoter (pGL3-NFAT-luc) and reference plasmids with Renilla luciferase (pRL-SV40). Negative controls were wild type PC12 cells transfected with promoterless pGL3-luc plasmids and positive controls were wild type PC12 transfected with plasmids overexpressing NFAT together with the pGL3-NFAT-luc (pGL3-NFAT-luc-NFAT+/+). NFAT activity was determined with a luciferase reporter dual assay (Thermo Scientific Pierce) and showed as the ratio of the luminescence signals derived from Firefly and Renilla luciferases. Bars represent mean values ± SEM. Symbols: control cells (C), PMCA2-deficient cells (_2), PMCA3-deficient cells (_3) and pGL3-NFAT-luc+/+NFAT – wild type cells overexpressing NFAT. Student’s t-test was used for comparison of control cells with PMCA2- or PMCA3-deficient cells. *P≤0.05, n = 5 (A). Nuclear content of dephosphorylated NFAT1 and NFAT3 was analyzed by immunoblotting. Protein bands were quantified densitometrically, standardized to PARP (nuclear marker) and normalized to control cells, expressed as y = 1. Bars represent mean values ± SEM. Student’s t-test was used for comparison of control cells with PMCA2- or PMCA3-deficient cells. *P≤0.05, n = 6 (B). Representative immunoblots of nuclear content of dephosphorylated NFAT1 (C) and NFAT3 (D) are demonstrated. Symbols correspond to: control cells (C), PMCA2-deficient cells (_2), PMCA3-deficient cells (_3).