Figure 6.
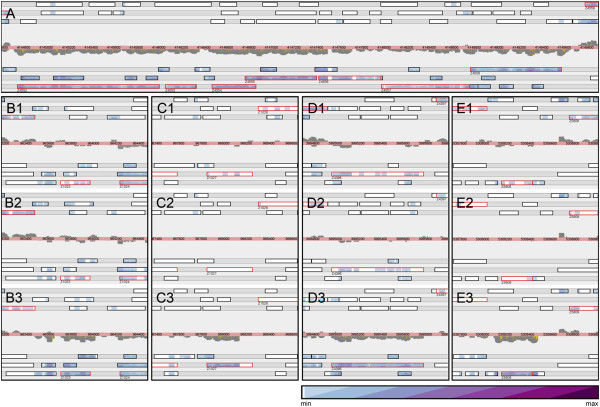
Visualization of the sequencing reads (= transcription) using the NGS overlap searcher [93]. The tool shows a plot of the read coverage in the middle. Forward strand reads are plotted above, reverse strand reads below the center line. The read starts are indicated in yellow. The bars above (forward strand) and below (reverse strand) the middle bar show all ORFs ≥ 90 nt in the six different reading frames. Annotated ORFs are in red. The tool shows the coverage also in the ORF bars according to the scaling in the lower right corner. A: selection of an “empty” region of the genome on the forward strand (4144776 – 4149762). The coverage shown is a sum signal of all eleven conditions sequenced on the SOLiD system. Only eight reads are found on the forward strand of this region. B1-B3: example for a regulated hypothetical gene, Z1023 (see Table 2), in LB medium (B1), minimal medium (B2), and on radish sprouts (B3). C1-C3: regulated hypothetical gene Z1027 (see Table 2), in LB medium (C1), minimal medium (C2), and on radish sprouts (C3). D1-D3: regulated hypothetical gene Z4396 (Table 2), in LB medium (D1), minimal medium (D2), and on radish sprouts (D3). E1-E3: regulated hypothetical gene Z5808 (see Table 2), in LB medium (E1), minimal medium (E2), and on radish sprouts (E3). The color map values range from 0.1 to 3 × 105, the exact expression values for each gene are listed in Table 2.
