Figure 4.
PRC1 Has a Genome-wide Role in PRC2 Recruitment and Polycomb Domain Formation at Target Sites in Mouse ESCs
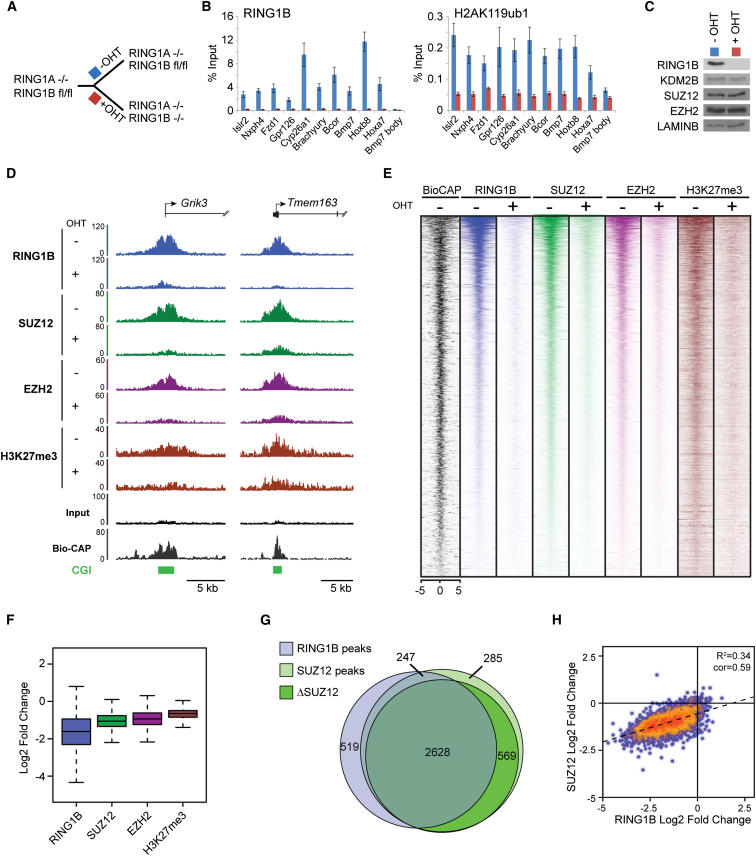
(A) A schematic of the Ring1a−/−Ring1bfl/fl system in which addition of tamoxifen (OHT) leads to deletion of RING1A/B.
(B) ChIP analysis for RING1B and H2AK119ub1 at polycomb target sites before (−OHT) and after 48 hr (+OHT) tamoxifen treatment. ChIP experiments were performed at least in biological duplicate with error bars showing SEM.
(C) Western blot analysis of polycomb factors following deletion of RING1A/B.
(D) Snapshots of ChIP-seq traces for RING1B, SUZ12, EZH2, and H3K27me3 in the Ring1a−/−Ring1bfl/fl cells prior to (−OHT) and following 48 hr +OHT treatment at the Grik3 and Tmem163 genes. Bio-CAP indicates nonmethylated DNA and CpG islands (CGI) are shown as green bars.
(E) Heat map analysis of SUZ12 peaks (n = 3,819), showing ChIP-seq data for RING1B, SUZ12, EZH2, and H3K27me3 at a 10 kb region centered over the SUZ12 peaks prior to (−OHT) and after 48 hr +OHT treatment. Bio-CAP is included to indicate nonmethylated DNA at these sites.
(F) A box and whisker plot showing log2 fold changes in normalized read counts comparing the ChIP-seq signal at SUZ12 sites −OHT and after 48 hr +OHT treatment.
(G) A Venn diagram showing the overlap of RING1B (light blue) and SUZ12 (light green) peaks including a further segregation of SUZ12-bound regions that show a greater than 1.5-fold change in SUZ12 occupancy (ΔSUZ12, dark green) after 48 hr tamoxifen treatment (+OHT).
(H) A scatter plot comparing the fold change of RING1B and SUZ12 at SUZ12 peaks −OHT and after 48 hr +OHT treatment.
See also Figures S2 and S3.