Figure S1.
Variant PRC1 Complex-Dependent PRC2 Targeting Occurs on an Inert TetO Array Template and at a Single Natural TetO Site in Mouse ESCs, Related to Figure 1
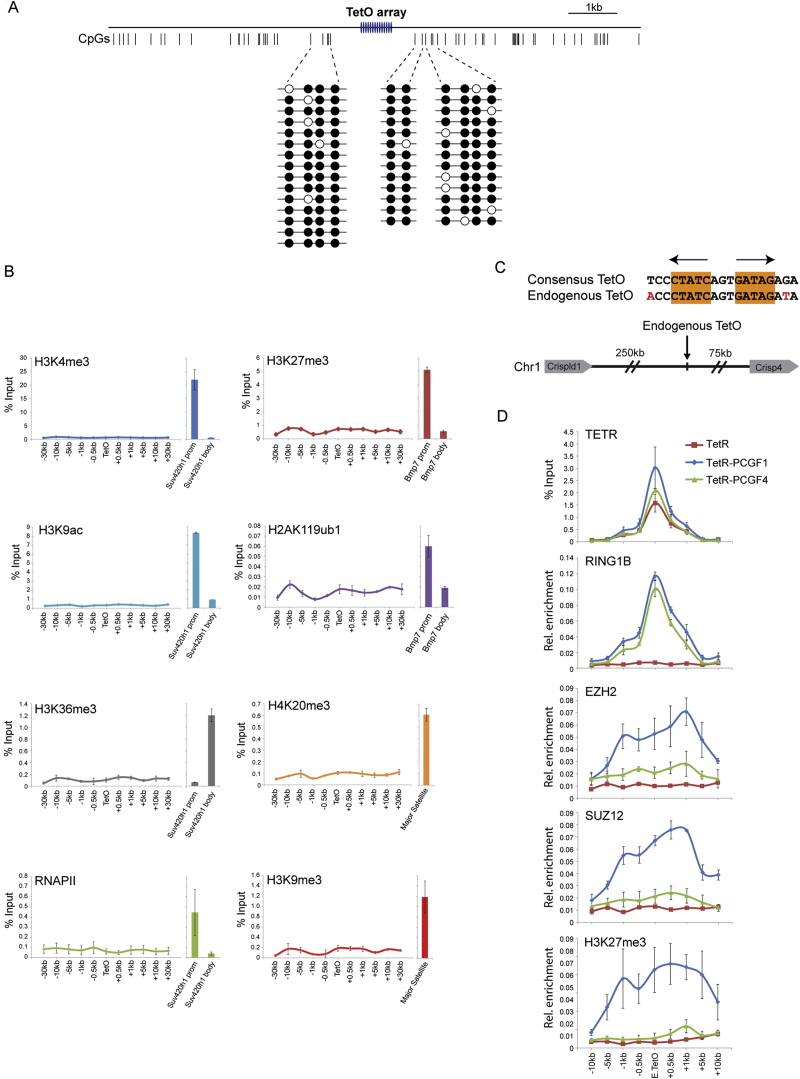
(A) Bisulfite sequencing reveals that CpG dinucleotides flanking the engineered TetO array in the Human BAC DNA remain methylated, as they are in human tissue, when stably inserted into the mouse genome. This indicates that normal DNA methylation features are recapitulated on the integrated DNA sequence.
(B) ChIP-qPCR analysis of the human BAC/TetO array stably integrated into mouse ESCs. ChIP was performed with antibodies specific for permissive chromatin marks and features of active transcription (left: H3K4me3, H3K9ac, H3K36me3, and RNAPII) and repressive chromatin features (right: H3K27me3, H2AK119ub1, H4K20me3, and H3K9me3). For each antibody, qPCR enrichment at a known target site is shown as a positive control. For gene-associated chromatin modifications this includes promoter (prom) and body amplicons for the indicated genes, while H3K9me3 and H4K20me3 were analyzed at sites in the repetitive major satellite regions. Together these observations indicate that the human BAC DNA, when inserted into mouse ESCs, retains its inert features and remains suitable for studying polycomb domain formation in tethering assays.
(C) A position on mouse chromosome 1, distant from surrounding genes, contains a single site that has a high degree of homology to a consensus bacterial TetO-binding site. This provides a “natural” site in the genome at which to analyze TetR fusion protein binding and function.
(D) ChIP-qPCR analysis of TetR, TetR-PCGF1 (variant PRC1), and TetR-PCGF4 (canonical PRC1) binding at the single natural TetO and flanking regions (Top). In each case, occupancy of PRC1 leads to RING1B recruitment (second panel). However, as observed at the engineered TetO array EZH2, SUZ12, and H3K27me3 are only recruited to the endogenous TetO site when a variant PRC1 complex occupies the site (bottom three panels). Together these observations demonstrate that variant PRC1 complex-dependent PRC2 recruitment and de novo polycomb domain formation is observed at a “natural” sequence in the mouse genome, and is not the result of copy number or site specific features inherent to the engineered TetO array. All ChIP experiments in (B and D) were performed in biological duplicate with error bars showing SEM.