Figure S2.
Conditional Deletion of RING1A/B Results in Depletion of H2AK119ub1, Loss of PRC2 Occupancy, and H3K27me3 at Target Genes Independently of the Magnitude of Associated Gene Expression Change, Related to Figure 4
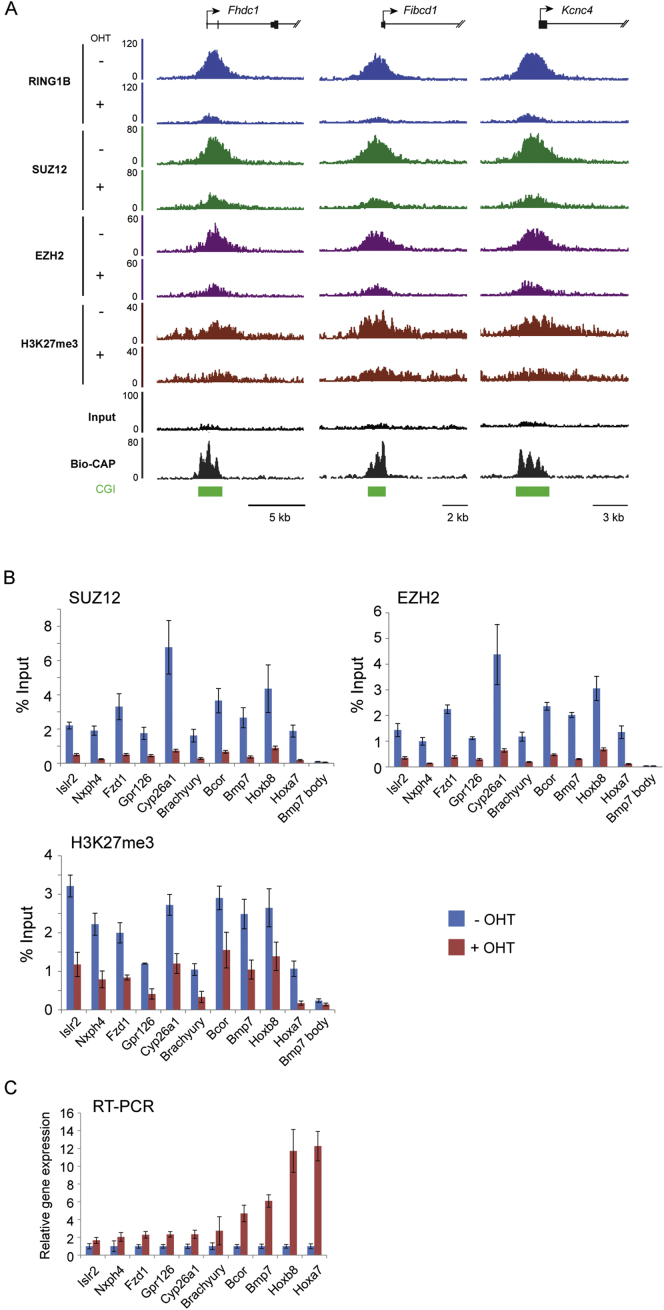
(A) Snapshots of ChIP-seq traces for RING1B, SUZ12, EZH2 and H3K27me3 in the Ring1a−/−Ring1bfl/fl cells prior to (−OHT) and following 48 hr (+OHT) of tamoxifen treatment. Three representative genes are depicted illustrating the reduction in RING1B, SUZ12, EZH2, and H3K27me3 following removal of the RING1A/B. A Bio-CAP sequencing trace is shown to indicate the location of nonmethylated DNA and a CpG Island (CGI) prediction annotation track is show as green bars under the traces. This complements examples already shown in main Figure 4.
(B) ChIP-qPCR analysis for PRC2 components SUZ12, EZH2, and H3K27me3 at a series of sites showing loss of these factors in the ChIP-seq analysis in main Figure 4 after tamoxifen treatment. All ChIP experiments in (C-D) were performed in biological triplicate with error bars showing SEM.
(C) Gene expression analysis for the polycomb target genes analyzed by ChIP in (B). There is no correlation between gene expression change and scale of polycomb group protein loss, suggesting that altered gene expression is not driving PRC2 loss. RT-PCR was performed in biological triplicate and is normalized to Hprt1 expression. Error bars show SEM.